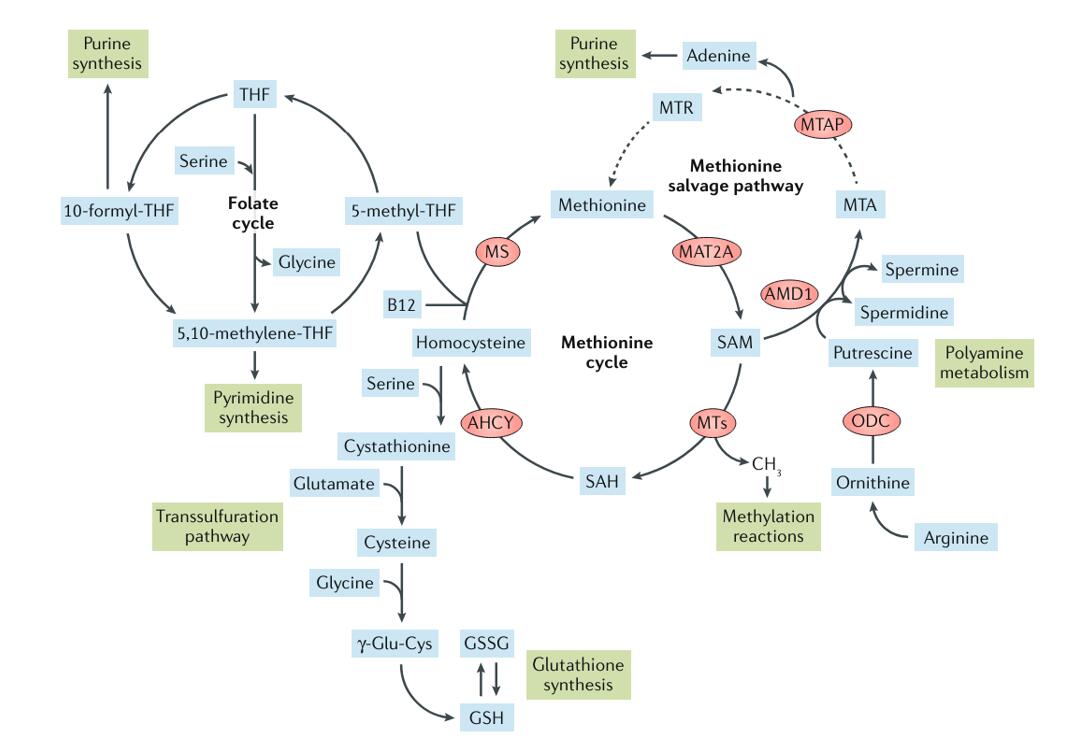
The methionine cycle, also known as the one-carbon metabolism pathway, is a fundamental biochemical pathway involved in the synthesis and recycling of the amino acid methionine and its derivative, S-adenosylmethionine (SAM). This cycle serves as a hub for various metabolic pathways, influencing cellular processes ranging from protein synthesis to DNA methylation.
Methionine Metabolism
Methionine Synthesis:
Methionine, an essential amino acid, cannot be synthesized by the body and must be obtained from the diet. The synthesis of methionine primarily occurs through a pathway known as the transmethylation cycle or methionine salvage pathway. In this pathway, homocysteine, an intermediate in methionine metabolism, is methylated to form methionine. This reaction is catalyzed by the enzyme methionine synthase, which utilizes methylcobalamin (a form of vitamin B12) as a cofactor. Methionine synthase transfers a methyl group from 5-methyltetrahydrofolate to homocysteine, yielding methionine and tetrahydrofolate.
Role in Protein Synthesis
Methionine plays a crucial role in protein synthesis as the initiator amino acid. During translation, the initiator tRNA (tRNAi^Met) binds to the start codon of the mRNA on the ribosome, carrying methionine. This initiation complex is essential for the accurate initiation of protein synthesis. Following initiation, methionine may undergo post-translational modifications, such as formylation or cleavage by methionine aminopeptidase, depending on the specific protein and cellular context.
S-Adenosylmethionine (SAM) Synthesis
A significant portion of methionine is utilized for the synthesis of S-adenosylmethionine (SAM), a universal methyl donor in numerous cellular methylation reactions. SAM is synthesized from methionine and ATP by the enzyme methionine adenosyltransferase (MAT). SAM serves as a methyl group donor for various methylation reactions, including DNA methylation, histone modification, and the methylation of proteins and lipids. These methylations play essential roles in regulating gene expression, chromatin structure, and cellular signaling pathways.
Transsulfuration Pathway
Alternatively, methionine can be metabolized via the transsulfuration pathway, leading to the synthesis of cysteine, another important amino acid. In this pathway, homocysteine is converted to cysteine through a series of enzymatic reactions. The first step involves the condensation of homocysteine with serine to form cystathionine, catalyzed by the enzyme cystathionine β-synthase (CBS). Cystathionine is then cleaved by cystathionine γ-lyase (CGL) to yield cysteine, α-ketobutyrate, and ammonia.
Regulation of Methionine Metabolism
The enzymes involved in methionine metabolism are tightly regulated to maintain cellular homeostasis. Regulation occurs at multiple levels, including transcriptional, post-transcriptional, and post-translational mechanisms. Additionally, enzyme activity can be modulated by allosteric regulation, covalent modification, and interactions with regulatory proteins. Nutrient availability, cellular energy status, and signaling pathways also influence the activity of enzymes within the methionine metabolic pathway, ensuring proper coordination with cellular metabolic processes.
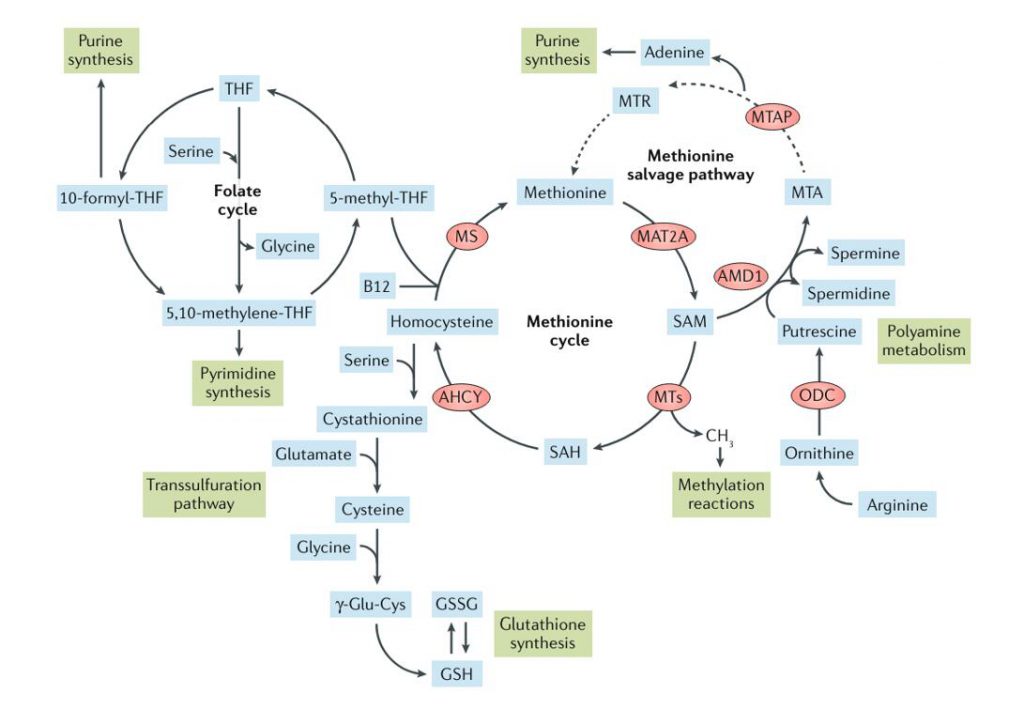
Methionine metabolism and related metabolic processes (Sanderson et al., 2019)
Interplay with Other Metabolic Pathways
Methionine metabolism intersects with various other metabolic pathways, including the folate cycle, the transsulfuration pathway, and the Krebs cycle. Metabolites from these pathways can feed into methionine metabolism or be derived from methionine metabolism. This interconnected network of metabolic pathways enables the integration of cellular metabolism and ensures the availability of essential metabolites for cellular function and homeostasis.
Key Enzymes and Reactions
Methionine Adenosyltransferase (MAT)
Enzyme Function: Methionine adenosyltransferase (MAT) is responsible for catalyzing the formation of S-adenosylmethionine (SAM) from methionine and ATP.
Reaction: Methionine + ATP → S-Adenosylmethionine (SAM) + Pi
Importance: SAM is a crucial metabolite serving as a universal methyl donor for numerous methylation reactions in the cell. These reactions include DNA methylation, histone modification, and methylation of proteins and lipids, which are essential for regulating gene expression, chromatin structure, and various cellular processes.
Regulation: MAT activity is regulated by various mechanisms to maintain SAM homeostasis. Feedback inhibition by SAM and other methylated metabolites serves as a mechanism for regulating SAM levels. Additionally, MAT expression can be modulated by transcriptional regulation in response to cellular signals and metabolic demands.
Methionine Synthase:
Enzyme Function: Methionine synthase catalyzes the conversion of homocysteine to methionine, using methylcobalamin (vitamin B12) as a cofactor.
Reaction: Homocysteine + Methylcobalamin → Methionine + Tetrahydrofolate
Importance: Methionine synthase plays a critical role in maintaining methionine levels for protein synthesis and SAM synthesis. By converting homocysteine to methionine, methionine synthase ensures the replenishment of methionine pools, essential for cellular growth, proliferation, and protein synthesis.
Regulation: The activity of methionine synthase is regulated by various factors, including the availability of methylcobalamin (vitamin B12) and folate derivatives. Deficiencies in these cofactors can impair methionine synthase activity, leading to elevated homocysteine levels and perturbations in methionine metabolism.
Methionine Adenosyltransferase Reductase (MTRR)
Enzyme Function: Methionine adenosyltransferase reductase (MTRR) participates in the regeneration of the active form of MAT, sustaining SAM synthesis.
Reaction: NADPH + Methionine Adenosyltransferase (MAT-III) → Methionine Adenosyltransferase (MAT-I) + NADP+
Importance: MTRR ensures the continuous synthesis of SAM by regenerating the active form of MAT, which catalyzes the formation of SAM from methionine and ATP. This enzyme plays a crucial role in maintaining SAM levels, thereby influencing methylation reactions and cellular processes dependent on SAM.
Regulation: MTRR activity is regulated by factors such as cellular redox status and availability of NADPH, which serve as cofactors for MTRR-mediated reduction of MAT.
Regulation of the Methionine Cycle
Control Mechanisms Governing Methionine Metabolism
The regulation of the methionine cycle is crucial for maintaining cellular homeostasis and ensuring proper functioning of essential metabolic processes. Regulation occurs at multiple levels, including transcriptional, post-transcriptional, and post-translational mechanisms.
Transcriptional Regulation: The expression of genes encoding enzymes involved in the methionine cycle can be modulated at the transcriptional level. Transcription factors, such as SREBP-1c (sterol regulatory element-binding protein 1c) and NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells), can regulate the expression of genes encoding key enzymes in methionine metabolism. For example, SREBP-1c can activate the transcription of MAT2A (encoding MAT α1 subunit), a key enzyme in SAM synthesis, in response to cellular lipid levels.
Post-Transcriptional Regulation: RNA processing and stability mechanisms can influence the abundance of mRNA transcripts encoding methionine cycle enzymes. RNA-binding proteins and non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), can regulate mRNA stability and translation efficiency. For instance, miR-34a has been shown to target MAT2A mRNA, leading to decreased MAT α1 subunit levels and reduced SAM synthesis in hepatocellular carcinoma cells.
Post-Translational Modification: Enzyme activity within the methionine cycle can be modulated by post-translational modifications, such as phosphorylation, acetylation, and ubiquitination. These modifications can alter enzyme activity, stability, and subcellular localization. For example, phosphorylation of MAT by protein kinase C (PKC) has been shown to increase MAT activity, leading to elevated SAM levels and enhanced methylation capacity in cancer cells.
Factors Influencing Enzyme Activity Within the Cycle
In addition to gene expression regulation, the activity of enzymes within the methionine cycle is influenced by various factors, including cellular metabolite levels, nutrient availability, and signaling pathways.
SAM Feedback Inhibition: SAM acts as a feedback inhibitor of MAT, the enzyme responsible for SAM synthesis. Elevated SAM levels inhibit MAT activity, thereby reducing SAM synthesis and maintaining SAM homeostasis. This feedback inhibition mechanism ensures tight regulation of SAM levels in response to cellular methyl donor requirements.
Nutrient Availability: Nutrient availability, particularly amino acids and vitamins involved in methionine metabolism, can influence the activity of enzymes within the methionine cycle. For example, deficiencies in vitamin B12 or folate can impair methionine synthase activity, leading to elevated homocysteine levels and perturbations in methionine metabolism.
Cellular Energy Status: Cellular energy status, as reflected by levels of adenosine nucleotides (ATP, ADP, AMP), can impact enzyme activity within the methionine cycle. ATP serves as a substrate for SAM synthesis by MAT, while ADP and AMP can allosterically regulate enzyme activity. Changes in cellular energy status can therefore modulate SAM synthesis and methylation capacity.
Cellular Signaling Pathways: Signaling pathways, such as the mTOR (mechanistic target of rapamycin) pathway and the AMPK (AMP-activated protein kinase) pathway, can influence methionine metabolism through regulation of gene expression, enzyme activity, and nutrient sensing. For example, mTOR signaling promotes SAM synthesis by upregulating the expression of MAT2A, while AMPK activation inhibits MAT activity, leading to reduced SAM levels and altered methylation capacity.
Connections Between the Methionine Cycle and Other Metabolic Pathways:
Folate Cycle:
The methionine cycle is intricately connected with the folate cycle, forming a metabolic network known as one-carbon metabolism. In the folate cycle, 5-methyltetrahydrofolate serves as a methyl donor for the conversion of homocysteine to methionine by methionine synthase. This reaction replenishes the pool of methionine for protein synthesis and SAM synthesis. Conversely, SAM serves as a methyl donor for the remethylation of tetrahydrofolate to form 5-methyltetrahydrofolate, closing the loop between the methionine cycle and the folate cycle.
Transsulfuration Pathway:
The methionine cycle intersects with the transsulfuration pathway, which is responsible for the synthesis of cysteine, a precursor for glutathione, an important antioxidant molecule. In the transsulfuration pathway, homocysteine is converted to cysteine through a series of enzymatic reactions catalyzed by cystathionine β-synthase (CBS) and cystathionine γ-lyase (CGL). Methionine serves as a precursor for cysteine synthesis, highlighting the interdependence between methionine metabolism and antioxidant defense mechanisms.
Krebs Cycle (Citric Acid Cycle):
The methionine cycle indirectly influences the Krebs cycle through its interplay with other metabolic pathways. SAM serves as a methyl donor for the synthesis of carnitine, a molecule involved in fatty acid transport into the mitochondria for β-oxidation. Additionally, methionine metabolism contributes to the synthesis of α-ketoglutarate, an intermediate in the Krebs cycle, through the transamination of methionine to form α-ketobutyrate and methionine sulfoxide.
Reference
Sanderson, Sydney M., et al. "Methionine metabolism in health and cancer: a nexus of diet and precision medicine." Nature Reviews Cancer 19.11 (2019): 625-637.