Introduction to KEGG Annotation
Why Use KEGG?
Key Features of KEGG Annotation
Tools for KEGG Annotation
KEGG Annotation Analysis Service at Creative Proteomics
Introduction to KEGG Annotation
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a world-renowned bioinformatics resource that has revolutionized the way researchers analyze and interpret biological data. Established in 1995, KEGG was developed to integrate genomic, chemical, and functional information into a unified platform, enabling a systems-level understanding of biological processes. It serves as a vital tool for deciphering the complex networks that govern cellular functions and interactions, such as metabolism, signaling pathways, and genetic information processing.
KEGG annotation is the process of assigning biological meaning to genes, proteins, or molecules by linking them to entries in the KEGG database. This includes mapping genes to pathways, associating them with orthologous groups, and analyzing their roles within modular biological systems. By providing a structured framework for interpreting high-throughput genomic data, KEGG annotation is essential for advancing research in genomics, transcriptomics, proteomics, and metabolomics.
What sets KEGG annotation apart is its focus on system-level insights. It doesn't just list functions of individual genes or proteins; instead, it shows how these components interact within larger biological systems. For example, a researcher studying a disease like cancer can use KEGG to identify which pathways are disrupted by genetic mutations, revealing potential targets for therapeutic intervention. Similarly, in environmental microbiology, KEGG annotation helps map the functional roles of microbial communities in ecosystems.
Through KEGG annotation, researchers can answer critical questions, such as:
- What are the roles of specific genes or proteins in cellular pathways?
- How do genetic variations influence metabolic or signaling networks?
- Which pathways are involved in specific diseases, and how can they be targeted?
Why Use KEGG?
KEGG offers unique advantages compared to other bioinformatics databases. While resources like Gene Ontology (GO) and UniProt provide gene-level functional information, KEGG distinguishes itself by focusing on system-level integration. Its visual representation of pathways enables researchers to see how individual genes and proteins interact within broader biological networks.
Moreover, KEGG excels in mapping metabolic pathways, disease pathways, and drug-target interactions, making it invaluable for translational research. For example, in drug discovery, KEGG's pathway maps can help researchers identify potential therapeutic targets and predict drug efficacy by simulating interactions within metabolic systems. This focus on pathways and system-wide analysis is a defining feature of KEGG, offering a holistic view that complements the granular insights provided by other databases.
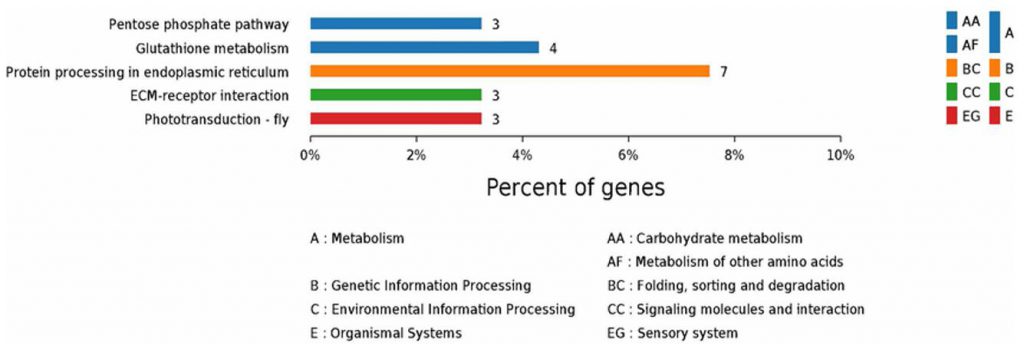
KEGG annotation of the top five pathways from the proteome database of the fed and unfed parasite groups (Li et al., 2019).
Key Features of KEGG Annotation
KEGG annotation stands out due to its comprehensive integration of biological, chemical, and functional data. This unique approach provides researchers with powerful tools to decode the complexities of life at a system-wide level. Let's dive deeper into the key features that make KEGG annotation an indispensable resource in bioinformatics.
Pathway-Centric Analysis
One of KEGG's most valuable features is its focus on pathways. KEGG PATHWAY maps depict biological processes as interconnected systems, highlighting the flow of information and metabolites within a cell. Unlike static lists of genes or proteins, these pathway maps allow researchers to visualize dynamic interactions and dependencies. Whether it's the glycolysis pathway, a signaling cascade, or a disease-specific pathway, KEGG provides an intuitive graphical interface to explore these intricate networks.
These maps are also interactive, enabling users to zoom in on specific reactions, molecules, or genes and access detailed annotations. This level of interactivity facilitates hypothesis generation, making KEGG PATHWAY a crucial tool for both exploratory research and targeted investigations.
Modular Approach with KEGG MODULE
KEGG MODULE is a feature that organizes genes and proteins into functional units, reflecting the modular nature of biological systems. Each module represents a specific biological process, such as ATP synthesis or nitrogen fixation. Unlike pathways, which often depict entire processes, modules focus on smaller, self-contained components that are easier to analyze and reconstruct.
For example, in metabolic engineering, researchers can use KEGG MODULE to identify and optimize specific enzyme sets required for desired chemical production. This modular framework is also ideal for studying incomplete datasets, such as metagenomic data, where not all genes in a pathway may be present.
Cross-Species Comparisons with KEGG Orthology (KO)
KEGG annotation leverages orthologous gene relationships to facilitate cross-species comparisons. KEGG ORTHOLOGY (KO) groups genes based on evolutionary relationships and shared functions, regardless of species. This allows researchers to transfer knowledge from well-studied organisms to less-characterized ones.
For instance, if a gene is identified in a newly sequenced plant genome, researchers can use KO to infer its function by comparing it to its orthologs in model organisms like Arabidopsis thaliana. This feature is particularly valuable in evolutionary biology, agricultural genomics, and comparative genomics.
Integration of Genomic, Chemical, and Environmental Data
KEGG seamlessly combines genomic data with chemical and environmental information. It includes databases like KEGG COMPOUND, KEGG GLYCAN, and KEGG DRUG, which link genes and pathways to biochemical molecules. This integration is essential for understanding how molecular components influence larger systems.
For example, KEGG COMPOUND provides detailed information about metabolites, enabling researchers to study metabolic flux and identify biomarkers. KEGG DRUG connects pathways to pharmaceuticals, aiding in drug repositioning and precision medicine.
Data Visualization and Usability
A standout feature of KEGG annotation is its emphasis on user-friendly visualization. The graphical pathway maps are designed to be both informative and aesthetically clear, making complex biological systems easier to understand. Additionally, tools like KEGG Mapper allow users to overlay their experimental data (e.g., gene expression or metabolomics) directly onto KEGG pathways, creating custom visualizations for presentations or publications.
Functional Reconstruction and Systems Biology Applications
KEGG annotation supports the reconstruction of entire biological systems, enabling researchers to move from individual genes to holistic models of cellular function. For example, KEGG can help reconstruct the metabolic network of an organism, even from incomplete genomic data, by filling gaps based on known modules and orthologs.
This feature is particularly useful for synthetic biology, where researchers design and engineer biological systems for specific purposes, such as biofuel production or disease modeling.
Tools for KEGG Annotation
KEGG Automatic Annotation Server (KAAS)
KAAS is an essential tool for high-throughput annotation of genomic and metagenomic datasets. It uses sequence similarity searches, typically BLAST or GHOSTX, to align query sequences against a set of reference gene datasets curated in KEGG. By employing a bidirectional best-hit (BBH) method, KAAS assigns KO (KEGG Orthology) identifiers to genes, enabling their placement in KEGG pathways, modules, and networks.
KAAS is particularly effective for de novo annotation of genomes, providing reliable functional predictions even for organisms without existing reference annotations. It is also used in metagenomic studies to infer functional potential from complex microbial communities. Its ability to automate annotation makes it ideal for large-scale projects requiring rapid yet accurate mapping.
KEGG Mapper
KEGG Mapper is a versatile tool designed to integrate user data with KEGG pathways and networks. It allows researchers to map genes, proteins, enzymes, or metabolites onto KEGG's curated pathway diagrams, facilitating the identification of active or disrupted pathways in biological systems. KEGG Mapper supports several functions, including:
- Pathway Mapping: Visualization of user data on KEGG pathways to identify trends such as upregulated or downregulated genes.
- Reconstruction of Pathway Maps: Enables users to reconstruct specific pathways based on custom input data, which is particularly valuable for comparative analyses between conditions.
- Module Completion Analysis: Assesses whether a set of genes or enzymes is sufficient to complete a functional module, aiding studies in metabolic engineering and synthetic biology.
By linking user data to functional pathways, KEGG Mapper provides actionable insights, such as pinpointing key enzymes in a metabolic bottleneck or identifying potential drug targets within disrupted pathways.
BlastKOALA and GhostKOALA
BlastKOALA and GhostKOALA are web-based tools developed as part of the KEGG platform, specializing in annotating smaller datasets or incomplete genomes. Both tools assign KO identifiers to genes based on sequence homology, but they differ in their underlying algorithms.
- BlastKOALA employs BLAST-based methods for precise annotation and is suitable for high-fidelity functional analyses.
- GhostKOALA utilizes a faster GHOSTX algorithm, making it ideal for large-scale or less complete datasets where speed is a priority.
These tools are particularly useful for researchers working on prokaryotic or eukaryotic genome projects that require accurate functional annotation within a short timeframe.
KEGG REST API
The KEGG REST API provides programmatic access to KEGG databases, allowing researchers to integrate KEGG annotations into custom pipelines or workflows. This flexibility is critical for large-scale bioinformatics studies that require automation or integration with other tools and databases. The API supports advanced queries, including retrieving pathway maps, extracting orthology groups, or analyzing metabolite interactions.
The API's capability to process data in bulk makes it a preferred choice for researchers conducting multi-omics analyses or comparative studies across multiple datasets. By leveraging KEGG REST API, bioinformatics workflows can be seamlessly scaled to accommodate complex datasets.
KEGG Annotation Analysis Service at Creative Proteomics
Creative Proteomics offers a comprehensive KEGG annotation analysis service, designed to help researchers uncover meaningful insights from complex genomic, proteomic, or metabolomic datasets.
Customized Solutions for Diverse Research Needs
Recognizing the unique requirements of different research projects, Creative Proteomics provides tailored solutions that accommodate a variety of data types and study objectives. Whether working on de novo genome annotation, metagenomic profiling, or multi-omics integration, the KEGG annotation service ensures accurate mapping of genes, proteins, or metabolites to KEGG pathways, orthology groups, and modules.
For example, in metagenomic studies, the service efficiently annotates microbial community functions, revealing critical insights into ecosystem dynamics or host-microbe interactions. In drug discovery projects, researchers benefit from pathway-level analysis to identify disease-specific biomarkers or druggable targets. This level of flexibility ensures that the service meets the demands of academic, clinical, and industrial research.
Key Benefits of the Service
- Time Efficiency: By outsourcing KEGG annotation tasks, researchers can focus on higher-level data interpretation and hypothesis generation without being burdened by the technical challenges of annotation workflows.
- High-Quality Data: The service guarantees robust and accurate annotations, minimizing errors that may arise from using automated pipelines alone.
- Scalability: Whether analyzing a single genome or a large metagenomic dataset, the service can handle projects of any scale.
- Comprehensive Reporting: Creative Proteomics provides detailed reports that include pathway visualizations, functional module analysis, and interpretive insights, helping researchers communicate their findings effectively.
Reference
Li, Yingdong, et al. "Comparative tandem mass tag-based quantitative proteomic analysis of Tachaea chinensis isopod during parasitism." Frontiers in Cellular and Infection Microbiology 9 (2019): 350.


