DIA Proteomics for FFPE and CSF: Robust Data from Fragile Clinical Samples
Formalin-fixed paraffin-embedded (FFPE) tissue blocks and cerebrospinal fluid (CSF) collections are some of the most valuable—and most difficult—clinical materials you can work with. A few sections from a decade-old tumor block, or a small CSF aliquot from a carefully phenotyped patient, may be all you ever get.
At the same time, these samples are messy:
- FFPE tissues are crosslinked, heterogeneous, and often old.
- CSF is low-protein, high dynamic range, and highly sensitive to pre-analytical variation.
If a proteomics run fails or produces unstable data, you cannot simply "repeat the experiment" with fresh material.
Data-independent acquisition (DIA) and 4D-DIA mass spectrometry have changed expectations here. DIA is now widely used to profile FFPE tissues and CSF with increased depth, fewer missing values, and better reproducibility, including in large clinical cohorts and neurodegenerative disease studies.
This resource focuses on a very practical question:
With FFPE and CSF that are precious, heterogeneous, and low input—is DIA proteomics really a robust option, and what does it take to make it work?
We'll walk through typical questions, technical realities, DIA-specific advantages, readiness checklists, and real-world use patterns, and how the NGPro™ platform is designed around these constraints.
Common Questions About FFPE and CSF DIA Proteomics
When we talk with clinical and translational teams about FFPE or CSF proteomics, the questions rarely start with "Which acquisition mode is best?". They sound more like this:
- "We only have 5–10 FFPE sections per patient. Is that enough for DIA proteomics?"
- "These FFPE blocks are 8–12 years old—do they still produce meaningful data?"
- "Our CSF aliquots are 200–300 μL, and a few have mild blood contamination—is the dataset still usable?"
- "Samples come from several hospitals with different fixation and handling—will batch effects swamp the biology?"
- "If this works, can we validate a small panel later without re-using all the archival material?"
Why FFPE and CSF Are So Challenging for Proteomics
FFPE: crosslinking, heterogeneity, and age
FFPE archives are a treasure for translational oncology and pathology: they encode tumor subtype, stage, treatment, and long-term outcomes in a form that can sit on a shelf for decades.
But for proteomics, FFPE introduces:
- Chemical crosslinking and modifications
Formalin fixation creates crosslinks and adducts that complicate protein extraction and digestion.
- Variable tissue content
Tumor cell content, necrosis, fibrosis, and immune infiltrates vary strongly between blocks and even within a single section.
- Block age and storage history
Older blocks can show increased oxidation and partial degradation, but are often those with the best clinical follow-up.
- Limited material
Needle biopsies and heavily used diagnostic blocks may yield only a handful of usable sections.
Despite these issues, multiple studies now show that carefully optimized workflows can generate thousands of quantified proteins from FFPE tissue—including ultra-low-input and spatial workflows.
CSF: low protein, high dynamic range, and pre-analytical sensitivity
CSF is a unique "liquid biopsy" for the central nervous system. It is particularly valuable in Alzheimer's disease, ALS, Parkinson's disease, and other neurodegenerative and inflammatory disorders.
The trade-offs:
- Low protein concentration and large dynamic range—few proteins dominate the signal, making low-abundance biomarkers hard to detect.
- Limited volume per patient, especially in research protocols where multiple assays share the same aliquots.
- Pre-analytical sensitivity
Delays in processing, centrifugation, or freezing, and even minor blood contamination, can affect the observed proteome.
DIA has emerged as a preferred strategy for CSF proteomics because it supports deeper coverage and more reproducible quantification from limited volumes, and is now used in ALS, AD, and multi-cohort neurodegenerative studies.
| Sample type | Key technical issues | Typical use cases |
| FFPE | Crosslinking, variable tumor content, variable block age, low input | Tumor subtyping, prognostic markers, IO response |
| CSF | Low protein, high dynamic range, blood contamination, limited volume | Neurodegeneration, neuroinflammation, drug response |
 Figure 1. Challenges of FFPE tissue and CSF for proteomics and how DIA/4D-DIA strategies address low input, heterogeneity and data completeness.
Figure 1. Challenges of FFPE tissue and CSF for proteomics and how DIA/4D-DIA strategies address low input, heterogeneity and data completeness.
Where DDA and TMT Fail on Heterogeneous, Low-Input FFPE and CSF Samples
DDA: strong discovery power, fragile on staged or multi-center cohorts
Data-dependent acquisition (DDA) selects the most intense precursors in each cycle for fragmentation. It's powerful for small discovery sets, but in FFPE and CSF projects it often struggles with:
- Stochastic sampling
Low-abundance or partially degraded peptides may be sampled in some runs but missed in others, leading to structured missingness. - Poor scalability
When you stitch together multiple centers or time windows, missing values and variable depth make downstream statistics unstable. - Limited re-use
Re-processing older DDA runs with new software can help, but stochastic precursor selection still limits matrix completeness.
TMT/iTRAQ: multiplexing at the cost of sample and design flexibility
Isobaric labeling methods are attractive when you can design everything up-front. In FFPE/CSF settings, reality is messier:
- Extra handling and protein loss
Every additional labeling and cleanup step is painful when you only have micrograms of protein. - Rigid batch design
Once plex layouts and bridge channels are chosen, adding new patients or centers later is difficult. - Multiple plexes for larger cohorts
Complex multi-batch normalisation is required, and a single labeling or instrument issue can compromise critical clinical samples.
For many FFPE and CSF projects, this leads to the same conclusion:
"We need something that tolerates low input, multi-center heterogeneity, and staged enrollment—without turning the design into a combinatorial nightmare."
How DIA and 4D-DIA Improve FFPE and CSF Proteomics Robustness
Systematic ion sampling and fewer missing values
In DIA, the instrument divides the m/z range into predefined windows and fragments all ions within each window, on every cycle. Instead of chasing only the top N precursors, DIA records a more complete and reproducible fragment-ion map of each run.
For FFPE and CSF, this translates into:
- More consistent sampling of low-abundance peptides and partially degraded material.
- Lower missingness at the protein level, which is crucial when comparing dozens or hundreds of clinical samples.
- Better suitability for regression, survival analysis, and multi-omics models, because the resulting protein × sample matrix behaves more like a quantitative feature table than a patchy discovery set.
Creative Proteomics's DIA Quantitative Proteomics Service is built around this principle, and explicitly supports FFPE, CSF, and other complex clinical matrices at cohort scale.
4D-DIA for low-input and complex clinical matrices
4D-DIA adds ion mobility separation as a fourth dimension to traditional LC-MS/MS (retention time, m/z, and intensity). This improves separation of co-eluting peptides and enables near-complete ion utilization, which is especially valuable for low-input samples.
For FFPE and CSF, 4D-DIA helps to:
- Increase sensitivity and depth at very low peptide loads.
- Reduce interference in complex backgrounds (e.g., plasma contamination in CSF, inflamed tumor microenvironment).
- Maintain high data completeness even in microsample or ultra-low-input workflows.
The 4D-DIA Quantitative Proteomics Services are specifically positioned for microsamples, large clinical cohorts, and high-throughput analysis, making them a natural fit for FFPE biopsies and limited CSF volumes.
FFPE- and CSF-specific DIA strategies
For FFPE tissue, DIA and 4D-DIA workflows need to accommodate crosslinking, partial degradation and variable tumor content. In practice, this means combining optimized deparaffinization, antigen retrieval, protein extraction and digestion with acquisition schemes that preserve sensitivity at low peptide loads.
For CSF, effective DIA proteomics typically relies on consistent pre-analytical handling, optional depletion or enrichment strategies, and DIA or 4D-DIA methods tuned for low total protein and a wide dynamic range, so that low-abundance biomarkers remain quantifiable.
Well-designed spectral library strategies—whether library-based or library-free—are critical for both matrices. Library-free DIA, in particular, has been successfully applied in CSF studies of ALS and other diseases, enabling re-analysis of raw data with updated tools and helping to refine biomarker candidates over time.
Readiness Checklists for FFPE and CSF DIA Projects
Even the best DIA pipeline can't fix fundamentally unsuitable input material. A quick readiness check with your pathologist or biobank team can save time and samples.
FFPE DIA readiness checklist
| Item | What to consider |
| Block age & storage | Age, storage conditions, known re-embedding or recuts |
| Section thickness & number | Typical thickness (e.g., 5–10 μm) and how many sections are available |
| Tumor content & necrosis | Estimated tumor cell percentage, necrosis, inflammatory infiltrate |
| Prior use | Has the block already been heavily sectioned or cored? |
| Clinical annotation | Diagnosis, stage, treatment, follow-up (for downstream modeling) |
CSF DIA readiness checklist
| Item | What to consider |
| Available volume | Volume per aliquot and number of aliquots per patient |
| Processing time | Delay from lumbar puncture to centrifugation and freezing |
| Freeze–thaw history | Number of freeze–thaw cycles planned or already used |
| Visible blood contamination | Hemolysis flag, RBC count, or visual inspection notes |
| Associated clinical data | Diagnosis, subtype, cognitive or functional scores, imaging |
If you're planning a large-scale FFPE or CSF campaign, Optimizing Sample Preparation for DIA Proteomics: Best Practices is a useful companion resource for pre-analytical planning.
NGPro™ DIA Workflow for Heterogeneous FFPE and CSF Samples
Instead of treating FFPE and CSF as "just another sample type", NGPro™ treats them as a distinct clinical asset with their own constraints.
Step 1 – Clinical material review and feasibility
The first step is not to set gradient length; it's to understand the material:
- Disease area (e.g., colorectal cancer, breast cancer, glioma, Alzheimer's, ALS).
- Sample sources and centers, block age distribution, CSF processing SOPs.
- Intended questions: diagnosis vs prognosis vs treatment response vs mechanism.
- Expected sample numbers and any planned validation cohorts.
This stage is where we help teams decide what is realistic: which blocks or CSF aliquots to prioritize, and whether standard DIA, 4D-DIA, or a mixed strategy makes sense.
Step 2 – Low-input optimized FFPE and CSF workflow with DIA / 4D-DIA
For FFPE, the FFPE Quantitative Proteomics Solutions include optimized deparaffinization, antigen retrieval, protein extraction, digestion, and QC tailored to archival tissue.
For CSF, the CSF Protein Quantitative Proteomics Solutions emphasize consistent processing, enrichment strategies when needed, and DIA/4D-DIA acquisition tuned for low abundance and high dynamic range.
Across both:
- DIA / 4D-DIA window schemes and gradients are chosen to balance sensitivity, depth, and throughput.
- QC samples (pools, references) monitor performance across batches, sites, and time.
- The same DIA-centric principles used in the Discovery Proteomics Service are applied, but with FFPE/CSF-specific optimizations.
Step 3 – From discovery signatures to robust panels
Discovery-scale DIA is often just the first pass:
- Broad FFPE or CSF profiling identifies candidate markers and pathways.
- Prioritized signatures are refined into short, robust panels suitable for larger validation cohorts or multi-center studies.
- The Targeted Proteomics Services (including DIA+PRM) then provide multiplexed, high-precision quantification of these panels in extended sample sets.
This "DIA for discovery, targeted for deployment" model is particularly important when each FFPE block or CSF sample is irreplaceable.
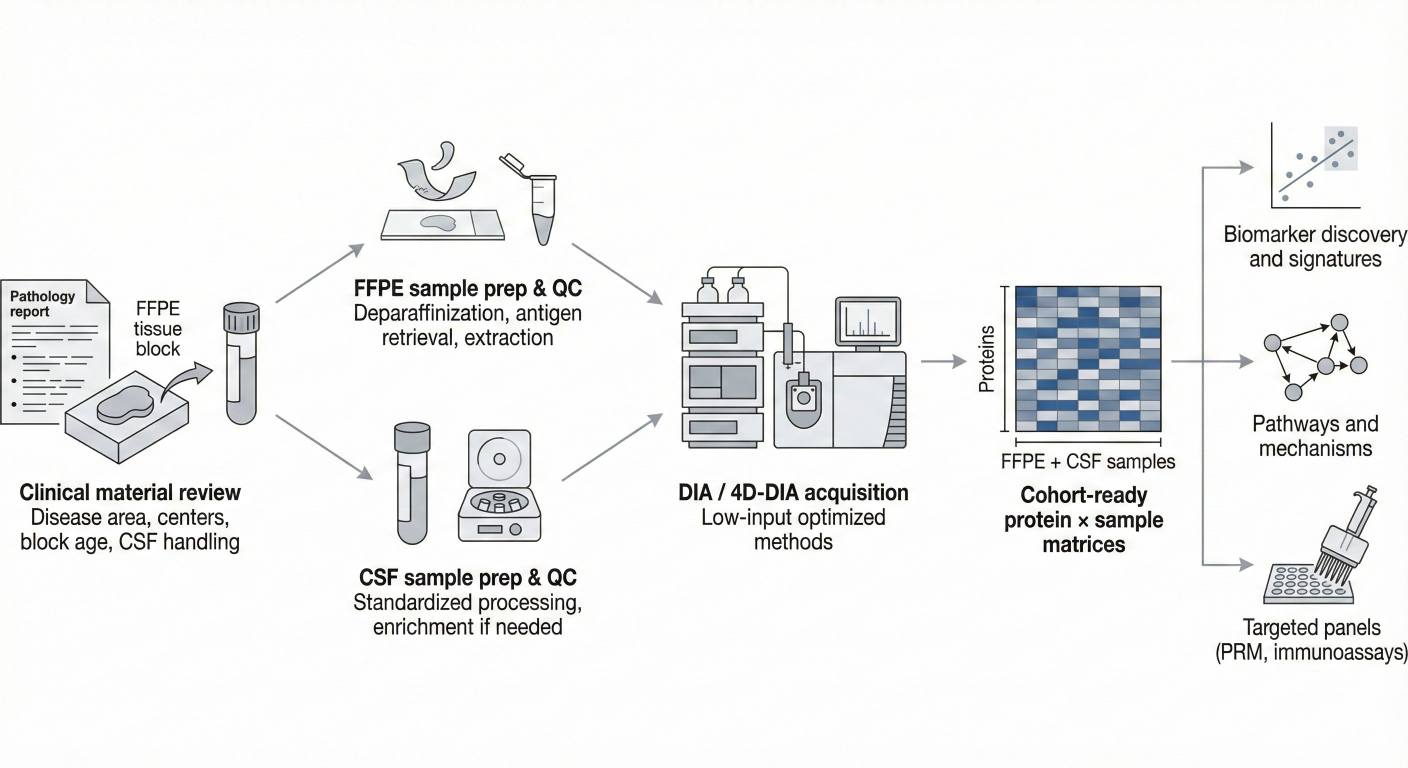 Figure 2. Low-input FFPE and CSF DIA workflow from clinical material review through sample preparation and DIA/4D-DIA acquisition to cohort-ready data and targeted panels.
Figure 2. Low-input FFPE and CSF DIA workflow from clinical material review through sample preparation and DIA/4D-DIA acquisition to cohort-ready data and targeted panels.
Use Cases and Expert Insights from FFPE and CSF DIA Projects
Below are example patterns that reflect how DIA proteomics is actually used in FFPE and CSF projects—combining published evidence with our team's experience.
1. Multi-center FFPE cohort for tumor subtyping and prognosis
Scenario
An oncology consortium collects ~220 FFPE tumor blocks from 4 hospitals for a specific cancer type. Blocks range from 2 to 12 years old, with heterogeneous fixation and variable tumor content. Long-term survival and treatment data are available.
Approach
- Pathology review stratifies blocks into "core discovery", "extended", and "low-priority" based on tumor percentage, necrosis, and block condition.
- DIA / 4D-DIA is applied to a carefully selected discovery subset, using harmonized extraction and QC across centers.
- Protein signatures are modeled against intrinsic subtypes and survival, following the logic of recent FFPE cohort studies showing high-throughput DIA workflows in archival tissue.
Expert view
- Do not send every block up-front; prioritize the combination of sample quality and clinical annotation.
- Plan in advance how a second wave (for validation) will be selected and processed using the same DIA pipeline, rather than "reinventing" the workflow later.
2. FFPE responders vs non-responders in immuno-oncology
Scenario
A translational group wants to understand why some patients respond to an immune checkpoint inhibitor while others do not. Only FFPE tumor samples are available; many blocks come from different centers.
Approach
- Use DIA to quantify immune-related pathways (antigen presentation, interferon signaling, T-cell trafficking) and microenvironment components from FFPE sections.
- Integrate proteomic signatures with PD-L1 staining, TMB, and clinical response categories.
- Borrow strategies from rectal cancer and other FFPE DIA studies that map treatment response with DIA-MS.
Expert view
- Equal attention must be given to pre-analysis FFPE QC and statistical modeling; otherwise, center-specific differences can masquerade as biology.
- A preplanned path from discovery signatures to targeted panels (via DIA+PRM) avoids repeated consumption of precious FFPE tissue during validation.
3. CSF DIA proteomics in neurodegenerative disease
Scenario
A neurology group has CSF from 150–200 participants, including patients with suspected Alzheimer's disease or ALS and age-matched controls. Volumes are modest, and there is no room for repeat experiments.
Approach
- Implement a DIA or 4D-DIA CSF workflow with standardized processing, monitoring hemolysis and other pre-analytical indicators.
- Quantify hundreds to thousands of proteins across samples and link them to clinical measures (cognitive scores, progression rates).
- Analyze data using network and module approaches similar to recent DIA CSF studies in ALS and AD that identify pathway-level signatures and refine biomarker candidates.
Expert view
- Invest in metadata discipline: time to processing, storage conditions, and basic QC flags matter as much as LC-MS settings when interpreting cohort-scale CSF data.
- Treat your DIA dataset as a long-lived resource; as new endpoints or algorithms emerge, re-analysis of the same raw files can unlock additional value without new lumbar punctures.
4. Combined FFPE + CSF signatures in translational pipelines
In some CNS tumor and neuroinflammatory programs, FFPE tumor tissue and CSF from the same patients are available:
- DIA on FFPE highlights tissue-level pathways and tumor microenvironment biology.
- DIA on CSF identifies circulating protein signatures related to barrier disruption, neuroinflammation, or neuronal injury.
Expert interpretation focuses on how these layers interact—for example, whether tissue pathways identified in FFPE translate into accessible CSF biomarkers that could support non-invasive monitoring.
When DIA Is – and Isn't – the Right Choice for FFPE and CSF
DIA proteomics is particularly well suited when:
- You need broad, unbiased coverage in FFPE or CSF across tens to hundreds of samples.
- You expect to reuse the same data as new endpoints or analysis tools appear.
- Multi-omics integration is planned, and you need a proteomics layer that behaves like a quantitative feature matrix.
Other approaches may be preferable when:
- You already have a small, fixed panel of proteins and only need ultra-high-throughput targeted assays.
- The question is very narrow and one-off, with no expectation of later re-analysis or cohort extension.
In many translational programs, the most pragmatic approach is blended:
- Use DIA (and 4D-DIA where needed) for discovery-scale profiling of selected FFPE and CSF cohorts.
- Translate robust signatures into targeted panels via the targeted proteomics.
- Reserve FFPE blocks and CSF aliquots strategically for future validation and orthogonal assays (IHC, ELISA, etc.).
For broader context on integrating proteomics into multi-omics pipelines, you may also find How to Integrate DIA Proteomics Data with Multi-Omics Platforms and Avoiding Failure in DIA Proteomics: Common Pitfalls and How to Fix Them helpful.
Frequently Asked Questions (FAQ)
References
- Lou, Ronghui, and Wenqing Shui. "Acquisition and analysis of DIA-based proteomic data: A comprehensive survey in 2023." Molecular & Cellular Proteomics 23.2 (2024): 100712.
- Weke, Kenneth, et al. "DIA-MS proteome analysis of formalin-fixed paraffin-embedded glioblastoma tissues." Analytica Chimica Acta 1204 (2022): 339695.
- Haines, Moe, et al. "High-throughput proteomic and phosphoproteomic analysis of formalin-fixed paraffin-embedded tissues." Molecular & Cellular Proteomics 24.9 (2025): 101044.
- Dellar, Elizabeth R., et al. "Data-independent acquisition proteomics of cerebrospinal fluid implicates endoplasmic reticulum and inflammatory mechanisms in amyotrophic lateral sclerosis." Journal of Neurochemistry 168.2 (2024): 115–127.
- Scalia, Elisabetta, et al. "Proteome profiling of cerebrospinal fluid and machine learning reveal protein classifiers of two forms of Alzheimer's disease characterized by increased or not altered levels of tau." Molecular & Cellular Proteomics 24.8 (2025): 101025.


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)