DIA Proteomics for Preclinical Studies: High-Throughput MoA and Off-Target Toxicity Screening
Preclinical pharmacology and toxicology teams are under pressure to answer harder questions with larger study designs than ever before.
Multiple candidates, multiple doses, multiple timepoints, multiple organs, plus replicates—very quickly a single in vivo study can generate 150–400+ samples. At that scale, traditional mass spectrometry workflows such as DDA and TMT/iTRAQ often become the bottleneck: workflows are complex, batch effects are difficult to control, and missing values undermine confident statistics.
Data-independent acquisition (DIA) mass spectrometry was built for this problem. By systematically sampling all ions within predefined windows, DIA delivers high-throughput, matrix-like datasets that are far better suited for large-scale preclinical studies and mechanism-of-action (MoA) profiling.
In this resource, we'll look at how preclinical teams can use DIA proteomics as a high-throughput engine for MoA and off-target toxicity screening—and how to implement an end-to-end workflow with the NGPro™ platform.
Why Preclinical Pharmacology and Toxicology Need High-Throughput Proteomics
From single markers to proteome-wide readouts
Traditional preclinical studies rely on:
- PK measurements
- A small set of PD markers
- Clinical chemistry
- Histopathology and functional scoring
These readouts are essential, but they rarely explain why a compound behaves the way it does. They provide limited insight into:
- Pathway-level activation or suppression
- Network-level rewiring across tissues
- Off-target pathway engagement that may predict later toxicity
Proteome-wide readouts from DIA mass spectrometry give a different level of visibility:
- Hundreds to thousands of proteins quantified per sample
- Pathway and network signatures across organs and timepoints
- Simultaneous MoA and early toxicity signal exploration within the same dataset
For compound ranking, hit-to-lead optimization, and early risk assessment, this level of resolution is increasingly valuable.
How preclinical study designs explode sample numbers
Many preclinical teams recognize the value of proteomics, but struggle with the scale of their designs. A typical study might include:
- 3–5 doses
- 3–6 timepoints
- 3–5 key organs (e.g., liver, kidney, heart, brain)
- 3–6 animals per group
Even with a modest design, this can easily reach 150–300+ samples. Multi-arm tox studies or multi-compound screens go higher still.
These are exactly the scenarios where search queries look like:
- "best proteomics method for >200 preclinical samples"
- "high-throughput proteomics for animal toxicity studies"
- "MoA proteomics screening in pharmacology models"
To support such scale, the proteomics workflow must be:
- High-throughput and robust
- Tolerant to complex matrices (tissues, plasma, CSF, etc.)
- Statistically friendly (low missingness, good reproducibility)
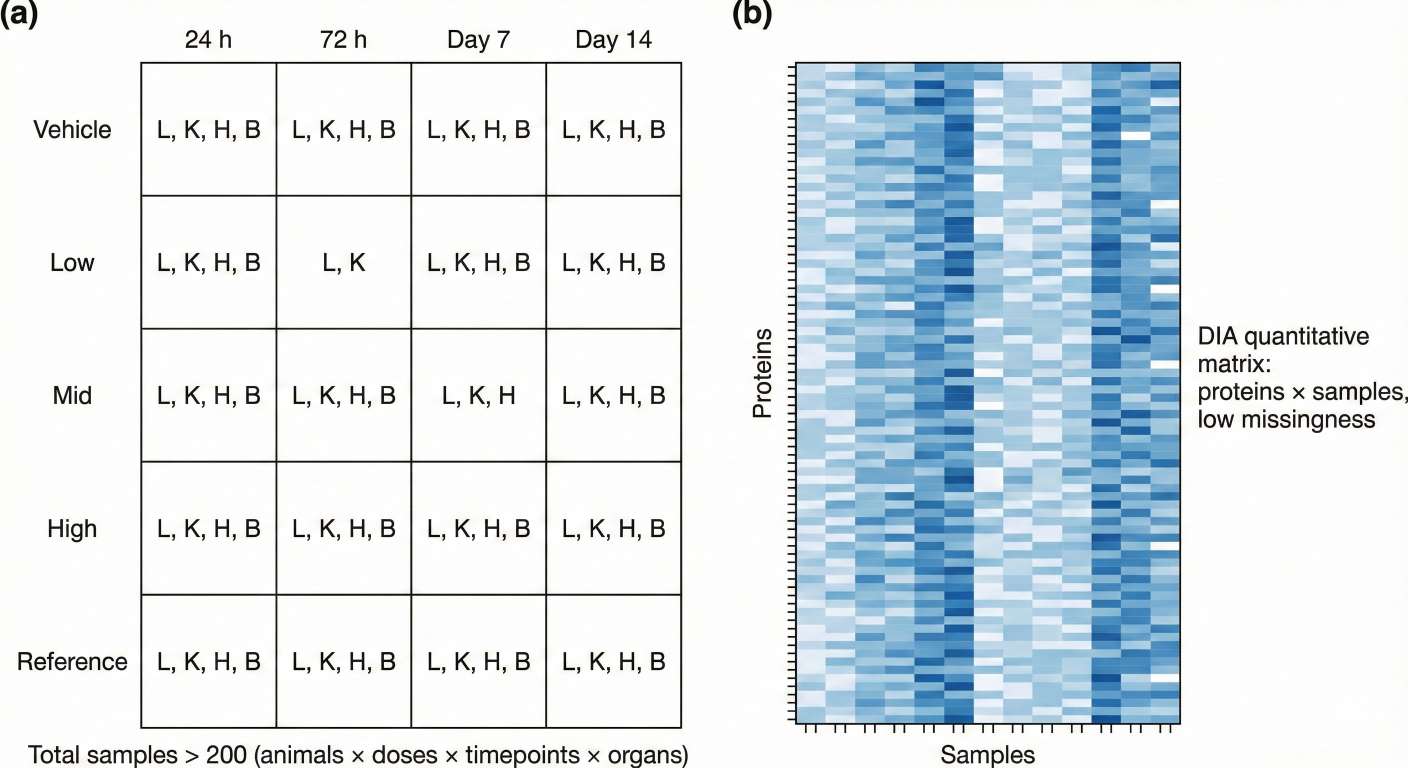 Figure 1. Large preclinical study design and resulting DIA proteomics data matrix.
Figure 1. Large preclinical study design and resulting DIA proteomics data matrix.
Why Classical DDA and TMT/iTRAQ Struggle in Large Preclinical Studies
Even when internal groups have access to LC-MS/MS, they often discover that standard DDA or multiplexed labeling workflows do not scale gracefully to large preclinical cohorts.
DDA: powerful depth, but unstable matrices for statistics
Data-dependent acquisition selects the most intense precursors for MS/MS in each cycle. That strategy is powerful for discovering new peptides, but in large cohorts it introduces challenges:
- Stochastic sampling: low-abundance peptides are not consistently selected across runs
- High missing values: especially problematic when comparing dose or time-response curves
- Run-to-run variability: makes it harder to detect subtle but biologically meaningful changes
For >100 samples across many conditions, this means:
- Incomplete data matrices
- Reduced statistical power
- Less confidence in MoA or tox signatures derived from DDA datasets
TMT/iTRAQ: complex workflows and limited scalability
Isobaric labeling strategies like TMT/iTRAQ offer multiplexing, but they are not always ideal for large preclinical studies:
- Complex experimental design: Balancing plex size, bridge channels, and batch-to-batch normalization.
- Ratio compression: Co-isolation and co-fragmentation can underestimate true fold-changes in complex samples.
- Scalability limits: 200–400 samples often require many multiplexed batches, increasing complexity and risk.
When a single labeling or instrument issue can compromise an entire batch, large animal studies become higher risk and harder to plan.
Method comparison at a glance
A simplified comparison for typical preclinical use cases is shown below:
| Method | Typical Strengths | Key Pain Points in >200-Sample Preclinical Studies |
| DDA label-free | Deep identification, good for small discovery sets | High missing values, stochastic sampling, weaker statistics across many runs |
| TMT/iTRAQ | Multiplexing, good within a single batch | Complex design, ratio compression, multi-batch stitching for large cohorts |
| DIA label-free | Systematic sampling, robust quantification | Requires dedicated analysis workflows, but scales well to hundreds of samples |
For teams facing large-scale pharmacology or toxicology studies, this is where DIA becomes particularly attractive.
If you want a deeper technical comparison between acquisition modes, you can refer to the resource: DIA vs DDA Mass Spectrometry: A Comprehensive Comparison.
DIA as a High-Throughput Engine for MoA and Toxicity Screening
Core principles in plain language
In DIA, the instrument does not "chase" only the most intense precursor ions. Instead, it:
- Divides the m/z range into predefined windows
- Cycles through those windows
- Fragments all ions in each window, every run
This strategy:
- Captures a complete and unbiased record of the sample's fragment ions
- Gives much more consistent sampling of low-abundance peptides
- Produces data that can be re-mined later as better libraries, algorithms, or biological questions emerge
For preclinical studies, the key benefits translate directly into study design advantages:
- Lower missing value rates
- Better run-to-run reproducibility
- Data matrices that are more amenable to statistical modeling and dose–response analyses
Why DIA is well suited to large preclinical cohorts
In large-scale proteomics research, DIA was explicitly developed to support robust quantification across many samples.
Applied to preclinical work, this means:
- High-throughput acquisition: modern platforms can process hundreds of samples with consistent methods
- Stable quantification: improved coefficients of variation and fewer missing values at the protein level
- Flexible downstream analysis: differential expression, clustering, and pathway analysis across doses, timepoints, and organs
DIA is also the core acquisition mode underpinning the NGPro™ discovery and targeted proteomics platform, which is designed for studies ranging from minimal sample numbers to large-scale cohorts.
4D-DIA and advanced DIA strategies
Next-generation DIA implementations add an ion-mobility dimension or refined window design to further enhance throughput and sensitivity. For example, 4D-DIA Quantitative Proteomics Services combine ion mobility separation with DIA scanning to deliver:
- Faster scanning with improved sensitivity
- Better separation of co-eluting peptides
- Higher data completeness, especially at low sample amounts
In practice, this allows preclinical studies to:
- Shorten gradients without sacrificing too much depth
- Maintain reliable quantification across many samples
- Explore more conditions (doses, timepoints, organs) within a fixed instrument budget
Study Designs: How to Use DIA Proteomics in Preclinical Drug Screening
DIA proteomics is flexible; it can support multiple study types with different emphases. Below are common design patterns used by pharmacology and toxicology teams.
MoA-focused studies in animal models
Typical questions
- How does my compound modulate signaling pathways compared with vehicle or reference drug?
- Are there proteomic signatures that distinguish responders from non-responders?
Example design elements
- Conditions: vehicle, multiple doses of test compound, optional reference drug
- Timepoints: early PD readouts (hours–days), mid-term efficacy windows
- Matrices: target organ tissue, plasma/serum, sometimes CSF or other fluids
- Readouts: pathway activation/inhibition, upstream/downstream components, network-level changes
DIA datasets from such designs can identify:
- Pathways that align with the intended MoA
- Compensatory or adaptive responses that may affect efficacy
- Potential biomarkers for later phase studies
Off-target and toxicity profiling
Typical questions
- Which organs show early proteomic signs of stress at sub-toxic or borderline doses?
- Are there pathway signatures that correlate with traditional tox findings?
Example design elements
- Conditions: vehicle vs multiple doses (including above therapeutic window)
- Timepoints: early (before histological damage), mid, and late stages
- Matrices: liver, kidney, heart, brain, and any organ with known risk profile
- Readouts: stress response pathways, mitochondrial function, inflammatory signatures, cell-death pathways
DIA allows preclinical teams to link early proteomic changes to:
- Later histopathology findings
- Clinical chemistry markers
- Functional deficits
This helps in building safety margins and in understanding off-target risks before entering clinical phases.
Dose–response and PK/PD integration
DIA proteomics also fits naturally with quantitative PK/PD thinking:
- Measure dose-dependent changes in pathway or protein signatures
- Overlay time-course proteomic data with exposure curves
- Identify exposure thresholds associated with desired or adverse pathway modulation
These design patterns benefit from stable quantitative performance across many samples, which is where DIA has clear advantages over purely DDA-based workflows.
End-to-End Workflow with the NGPro™ Platform
An effective preclinical DIA project is not just about the acquisition mode. It depends on a coordinated workflow from study design through data interpretation.
1. Pre-study consulting and experimental design
- Clarify biological objectives: MoA, toxicity, biomarker discovery, or combination
- Align on sample numbers, organs, doses, and timepoints
- Choose between DIA and 4D-DIA modes based on throughput and depth requirements
- Discuss potential downstream analyses (e.g., pathway modeling, clustering, integration with RNA-seq)
Early alignment ensures that the proteomics readouts will map cleanly onto decision points in the preclinical program.
2. Sample preparation and QC for animal tissues and biofluids
Preclinical samples often include:
- Frozen tissues (liver, kidney, heart, brain, muscle, etc.)
- Plasma/serum or other biofluids
- Occasionally CSF or specialized matrices
Standardized sample preparation and QC are critical to avoid avoidable variability.
Typical steps include:
- Verification of sample integrity and labeling
- Assessment of protein yield and extract quality
- Test runs or pilot gradients when appropriate
3. DIA or 4D-DIA acquisition optimized for throughput
Based on the agreed study design:
- LC gradients are selected to balance proteome depth with run time
- Instrument stability is monitored with QC injections and reference samples
- Acquisition parameters are standardized to maintain consistent performance across the entire cohort
For high-throughput studies where both depth and speed are critical, 4D proteomics services can further strengthen sensitivity and quantitative completeness by leveraging the ion-mobility dimension along with DIA.
4. DIA data analysis: from matrices to mechanisms
Raw DIA data require specialized processing; this is where robust pipelines and experience matter. The Data Independent Acquisition (DIA) Data Analysis service uses both spectrum-centric and peptide-centric strategies for:
- Signal processing, peak picking, and identification
- Normalization and batch assessment
- Differential protein expression across conditions
- Pathway and network analyses (e.g., enrichment, clustering, correlation with phenotypes)
Preclinical-specific outputs often include:
- MoA signatures distinguishing compound vs control or vs reference drug
- Organ-specific tox signatures that can be compared with histopathology
- Priority candidates (targets, markers, pathways) for follow-up validation
5. Target verification with DIA+PRM
Discovery-scale DIA is often followed by targeted validation on a smaller protein panel. The DIA+PRM Target Proteomics Services combine broad DIA discovery with highly specific PRM quantification:
- Translate a long candidate list into a focused set of markers
- Confirm dose-dependent behavior of key proteins across additional samples
- Build targeted assays that can later be used in more extensive toxicology or translational studies
This "DIA for discovery, PRM for confirmation" logic is especially appealing in preclinical workflows where multiple study phases reuse the same candidate markers.
If you're interested in how DIA-based proteomics integrates into broader multi-omics designs, you may also find How to Integrate DIA-MS Proteomics With Your RNA-seq Study helpful.
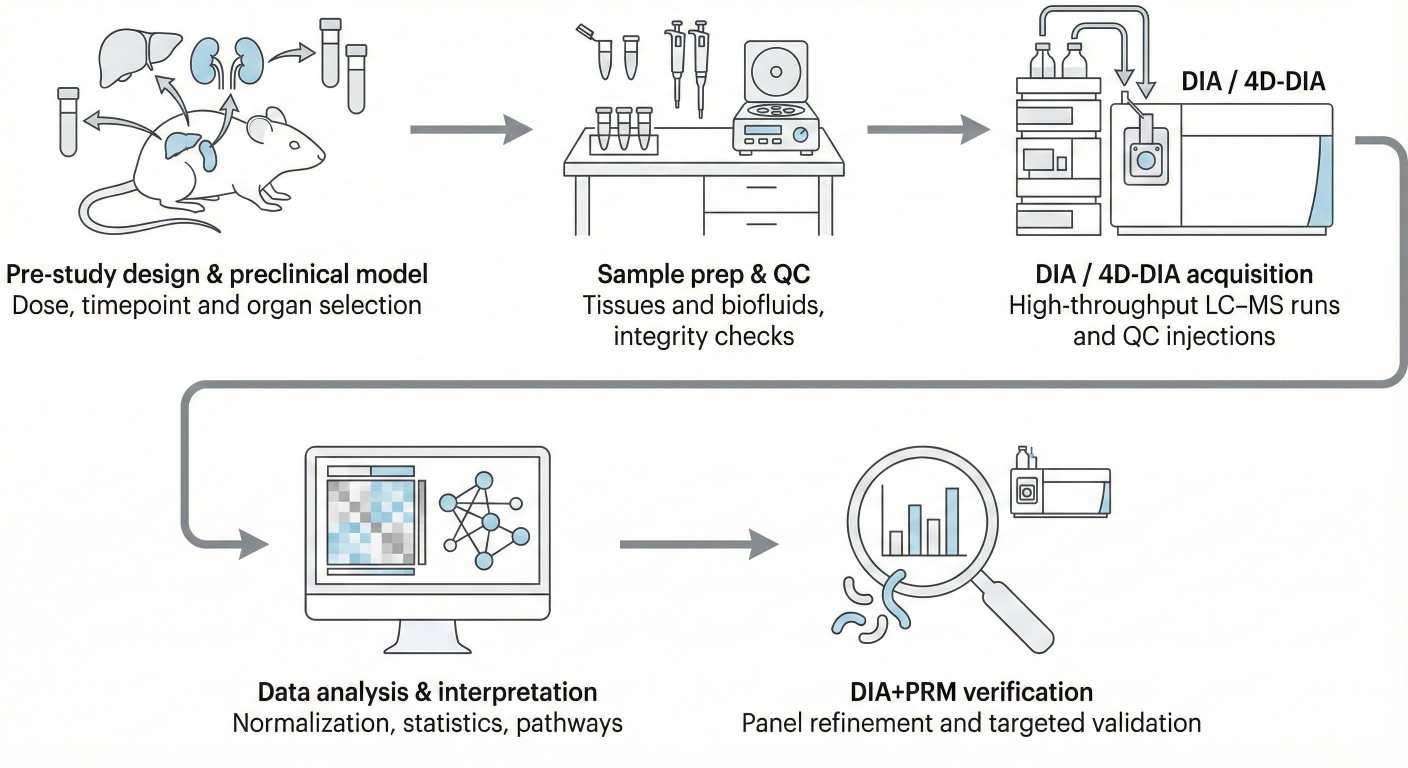 Figure 2. NGPro™ workflow for preclinical DIA proteomics from design to target verification.
Figure 2. NGPro™ workflow for preclinical DIA proteomics from design to target verification.
How Different Teams Use DIA Proteomics in Preclinical Projects
Different organizations apply the same core DIA principles to different business realities.
Small biotech: prioritizing leads with MoA-based signatures
- Situation: several promising hits, limited budget, and a tight timeline
- Approach: run a focused in vivo study on a subset of doses and timepoints, using DIA to generate pathway-level signatures
- Outcome: prioritize compounds that best align with desired MoA and show minimal early tox signatures, before committing to larger GLP programs
Large pharma: high-density tox and safety studies
- Situation: extensive dose-range finding, chronic exposure models, and multi-organ sampling
- Approach: integrate DIA proteomics into key tox studies to characterize organ-specific responses and early stress pathways
- Outcome: improved mechanistic understanding of adverse events, better linkage between exposure, histology, and molecular changes, and stronger internal confidence in candidate selection
Academic and translational labs: deep mechanism with limited resources
- Situation: one or two compounds or biologics of interest, but many timepoints or tissues
- Approach: use DIA-based discovery proteomics to maximize information yield per animal
- Outcome: richer mechanistic publications, better positioning for collaborations with industry, and datasets that can be re-mined as new hypotheses emerge
When DIA Is the Right Choice—and When to Consider Other Approaches
DIA is not the only option for all preclinical questions, but there are clear patterns where it is particularly strong.
DIA is especially well suited when:
- Sample numbers are moderate to large (dozens to hundreds)
- You need broad, unbiased molecular insight into MoA or toxicity
- Statistical modeling and clustering across conditions are important
- You may want to reuse the same dataset later for new hypotheses
Other approaches may be appropriate when:
- You only need to quantify a small, well-defined set of proteins in very large cohorts
- Regulatory or operational constraints favor established targeted assays
In many programs, a staged strategy works best:
- DIA-based discovery in representative preclinical models
- Selection of robust candidate markers or signatures
- Follow-up with targeted methods such as PRM or immunoassays in extended cohorts
The NGPro platform is designed to support this entire path, from early discovery to targeted validation.
Frequently Asked Questions (FAQs)
Quality and consistency come from a combination of standardized sample preparation, well-defined LC-MS methods, and rigorous data analysis. DIA workflows typically include:
- Internal reference or QC samples across batches
- Monitoring of system performance metrics
- Normalization strategies to correct minor technical variation
On the analysis side, spectrum-centric and peptide-centric strategies, such as those used in dedicated DIA data analysis services, help maintain depth and accuracy even when thousands of proteins are quantified.
Yes. One of the strengths of DIA is that it generates a dense, quantitative "matrix" of protein features that can be correlated with pharmacokinetic profiles, transcriptomic data, histology scores, or clinical chemistry parameters. In practice, this may involve:
- Aligning timepoints between assays
- Using shared IDs (animal, organ, treatment) across datasets
- Applying multivariate methods to link molecular signatures with phenotypic outcomes
This makes DIA proteomics a powerful bridge between molecular mechanism and in vivo pharmacology.
References
- Lou, Ronghui, and Wenqing Shui. "Acquisition and analysis of DIA-based proteomic data: A comprehensive survey in 2023." Molecular & Cellular Proteomics 23.2 (2024): 100712.
- Fröhlich, Klemens, et al. "Data-independent acquisition: A milestone and prospect in clinical mass spectrometry–based proteomics." Molecular & Cellular Proteomics 23.8 (2024): 100800.
- Bruderer, Roland, et al. "Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues." Molecular & Cellular Proteomics 14.5 (2015): 1400–1410.
- Song, Sufei, et al. "Quantitative proteomics analysis based on data-independent acquisition reveals the effect of Shenling Baizhu powder (SLP) on protein expression in MAFLD rat liver tissue." Clinical Proteomics 20.1 (2023): 55.
- Wang, Qiqi, et al. "Spike-in proteome enhances data-independent acquisition for thermal proteome profiling." Analytical Chemistry 96.49 (2024): 19695–19705.




 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)