Quantitative Profiling of Secreted Proteins for Cell Communication and Biomarker Discovery
NTA Service Advantages Workflow and Platform Sample Requirements Deliverables How to ChooseFAQ Get a Custom Proposal
What Is NTA-Based Exosome Identification
Nanoparticle Tracking Analysis (NTA) is a high-resolution technique for the characterization of extracellular vesicles (EVs), including exosomes, based on particle-by-particle tracking in liquid suspension. It enables absolute quantification and size distribution analysis, offering critical insights for R&D, formulation development, and process monitoring.
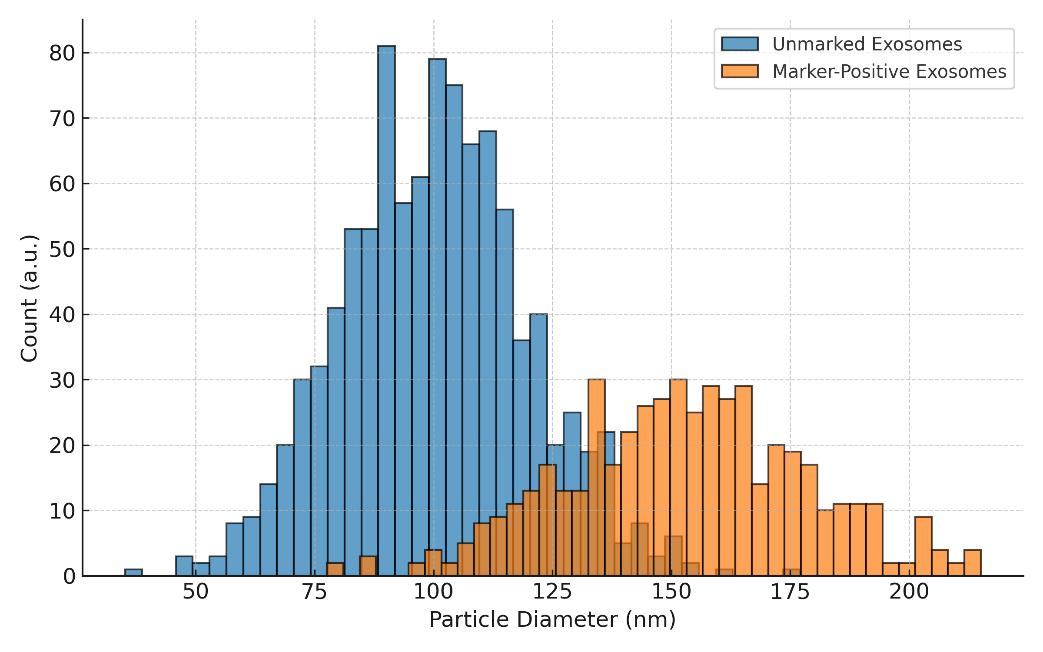
Creative Proteomics leverages NTA technology to deliver actionable metrics such as particle concentration (particles/mL), modal diameter, D10/D50/D90, and—via fluorescence labeling—marker-specific subpopulation detection (e.g., CD9⁺/CD63⁺/CD81⁺ EVs). This makes NTA especially valuable for users validating isolation methods, assessing sample integrity, or benchmarking EV-enriched preparations.
How Nanoparticle Tracking Analysis Works in Exosome Profiling
NTA is based on the principle of Brownian motion, where particles suspended in a fluid exhibit random motion. A laser beam illuminates the sample, and a sensitive camera captures the light scattered by each particle as it moves within a defined observation volume.
The software tracks multiple particle trajectories in real time and calculates their hydrodynamic diameter using the Stokes-Einstein equation, which relates particle displacement to size. This enables simultaneous sizing and counting of individual particles with high precision.
Because NTA analyzes particles individually, it can resolve polydisperse and heterogeneous populations, which are common in EV preparations. Two operational modes are typically used for exosome analysis:
- Brightfield (Conventional) NTA – Measures size and concentration of all detectable particles.
- Fluorescence NTA (fl-NTA) – Tracks only particles labeled with fluorescent markers, allowing for surface-marker-specific profiling (e.g., CD63⁺ vesicles).
What's Included in Our Exosome NTA Service
Particle Size and Concentration Analysis (Brightfield NTA)
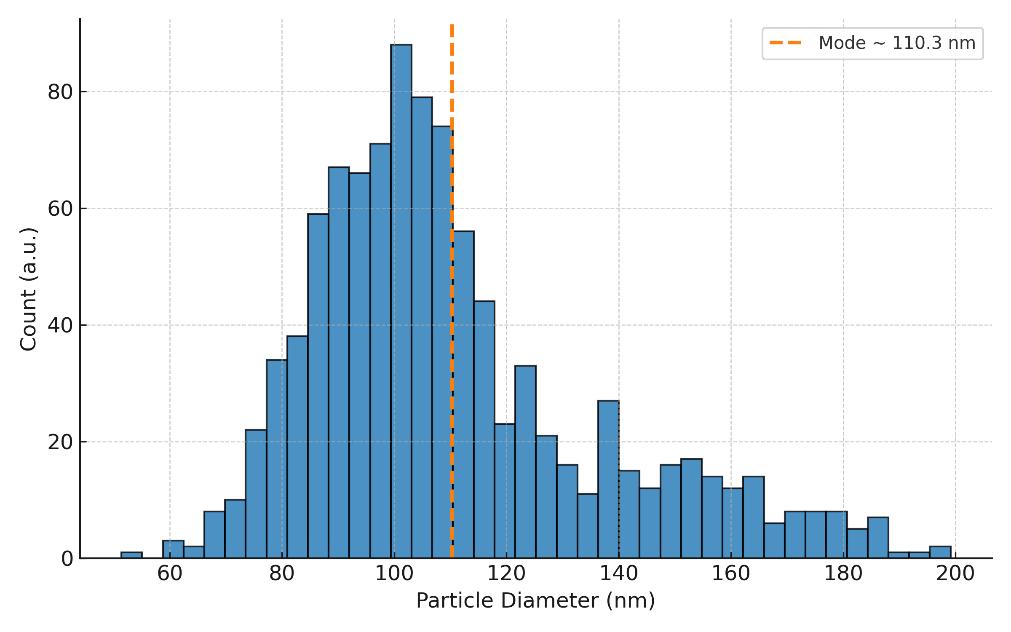
- Measures hydrodynamic diameter (mean, mode, D10/D50/D90)
- Provides absolute particle concentration (particles/mL)
- Generates size distribution histograms
- Performed with multiple replicates for statistical reliability
Fluorescent NTA for Marker-Positive EV Subsets
- Detects CD9⁺, CD63⁺, CD81⁺ or other fluorescently labeled exosome populations
- Compatible with multiple laser channels (405, 488, 532, 640 nm)
- Supports antibody or dye-based labeling
- Includes negative/isotype control validation
- Delivers subpopulation-specific size and count metrics
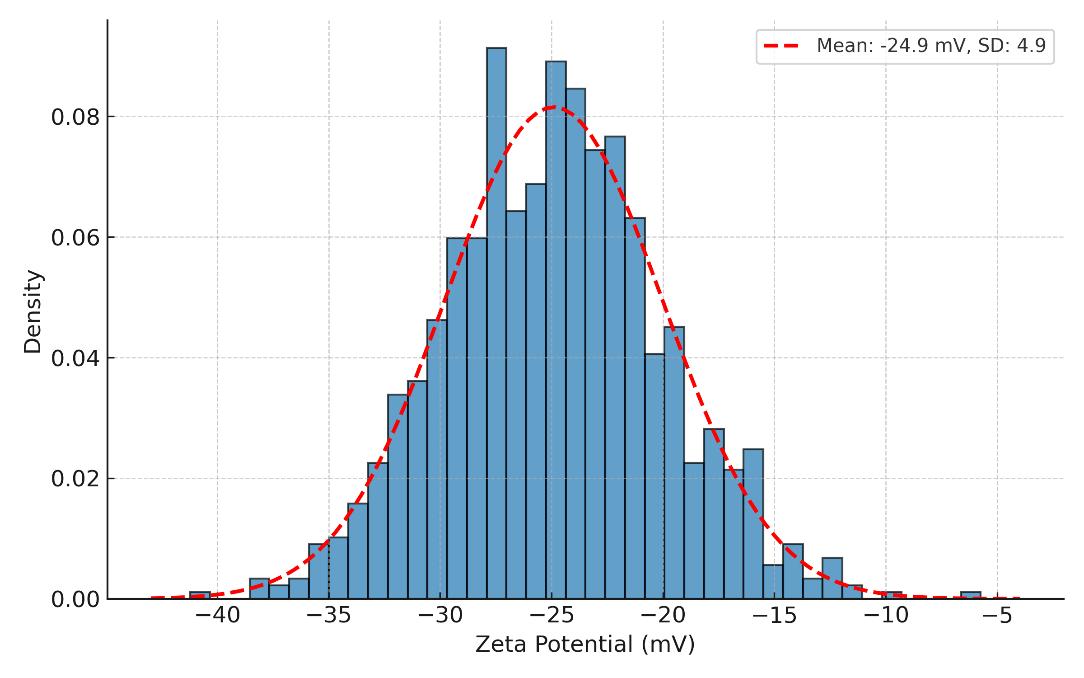
Zeta Potential Measurement
- Evaluates surface charge (ζ-potential) of exosomes
- Useful for stability assessment, formulation screening, and colloidal behavior profiling
Advantages of Our NTA Service
High-Precision Quantification
Accurate exosome concentration reporting with ±10% variance across replicates, ensuring reliable batch-to-batch comparison.
Validated Replicability
Every sample is measured in triplicate video recordings with strict QC checks, reducing variability and enhancing reproducibility.
Single-Particle Resolution
Detects particles as small as 30 nm, capturing size heterogeneity that bulk techniques like DLS cannot resolve.
Calibrated and Standardized Workflow
Instruments calibrated with traceable nanoparticle standards ensure consistent sizing and concentration results across different runs and projects.
Fluorescence-Enabled Specificity
fNTA supports up to 4 excitation channels (405/488/532/640 nm) for simultaneous detection of marker-positive exosomes (e.g., CD9, CD63, CD81).
Decision-Ready Data Packages
Clients receive publication-quality plots, structured datasets (CSV/JSON), and optional raw videos, making results integration-ready for R&D, QC, or publication.
Exosome Characterization Workflow Using Nanoparticle Tracking Analysis
1
Sample intake & prep
Confirm matrix/isolation/markers; pre-clarify and 0.22 µm filter; set dilution window.
4
Fluorescent NTA (optional)
Antibody/dye labeling; 405/488/532/640 nm channels; negative/isotype controls; marker-positive counts and size profiles.
2
Calibration & parameter lock
Traceable bead standards; lock camera level, detection threshold, temperature, and viscosity correction.
5
Zeta potential (optional)
Surface charge (ζ-potential) to inform stability and formulation decisions.
3
Brightfield NTA acquisition
Multi-video replicates; Brownian tracking to report concentration (particles/mL), mode, D10/D50/D90, and size histograms.
6
QC & reporting
Replicate agreement, dilution validity, background/artifact control; deliver PDF + CSV/JSON (raw videos/plots on request).