Quantitative Profiling of Secreted Proteins for Cell Communication and Biomarker Discovery
Proteomics Platforms Solutions by Goal Service Highlights Workflow Sample Requirements Deliverables FAQ Get a Custom Proposal
What Is Proteomics—and Why It Matters
Proteomics is the large-scale study of proteins, their expression levels, modifications, interactions, and functional roles across biological systems. As the key effectors of cellular activity, proteins provide dynamic insight into phenotype regulation, signaling networks, and disease mechanisms. Compared to genomics or transcriptomics, proteomics directly reflects the current physiological state of cells, tissues, or organisms.
Creative Proteomics delivers end-to-end proteomics solutions tailored for research in drug discovery, biomarker development, pathway elucidation, and more. Our platform integrates high-resolution mass spectrometry, advanced fractionation strategies, and AI-assisted bioinformatics, enabling quantitative and qualitative profiling across nearly all proteoforms.
Proteomics Technology Platforms
Flexible solutions for deep discovery, high-throughput screening, organelle mapping, and targeted validation.
Discovery Proteomics
Platform: Orbitrap Exploris 480 + nanoLC
This is our go-to platform for in-depth, unbiased proteome profiling. Equipped with high-resolution MS and low-femtomole detection sensitivity, it captures >10,000 proteins per sample and is ideal for:
- Differential expression across conditions or disease models
- Post-translational modification (PTM) screening
- Pathway mapping in early-stage discovery
With advanced label-free or TMT-based workflows, this platform supports comprehensive proteomic exploration from whole-cell lysates, tissues, or biofluids.
High-Throughput Proteomics
Platform: timsTOF Pro + Evosep One
Optimized for large sample cohorts, this configuration combines fast chromatographic gradients with trapped ion mobility separation (TIMS) for high-resolution, rapid quantitation. Perfect for:
- Biomarker screening in clinical or population-scale studies
- Longitudinal proteomic monitoring (e.g., time-course, treatment response)
- Efficient processing of >100 samples per week
Our pipeline ensures inter-batch consistency and minimal instrument downtime, making it ideal for large-scale comparative studies.
Organelle Proteomics
Platform: Q Exactive HF-X + proprietary fractionation kits
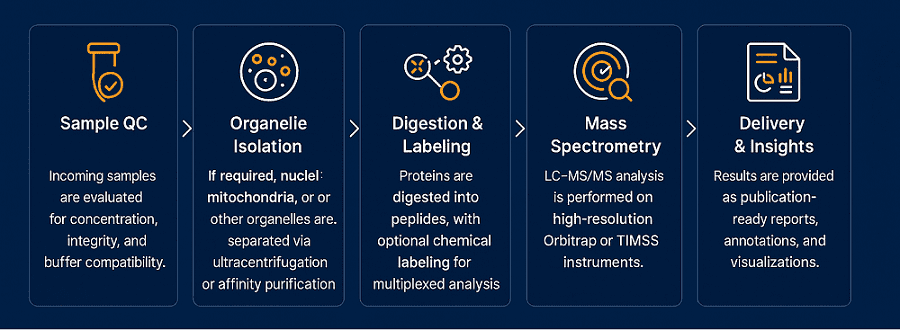
When spatial context matters, this platform enables precise localization of proteins within specific organelles. Using differential centrifugation, affinity purification, and contamination-reducing workflows, we can cleanly isolate:
This approach is critical for studying compartmentalized signaling, organelle-specific PTMs, or protein trafficking. Coupled with subcellular annotation algorithms, it delivers a spatially resolved proteomic landscape.
Targeted Quant Proteomics
Platform: QTRAP 6500+ / TripleTOF 6600
For clients requiring precise quantification of predefined protein panels, our targeted proteomics platform offers absolute quantitation using:
- MRM/SRM (Multiple Reaction Monitoring)
- PRM (Parallel Reaction Monitoring)
Ideal for:
- Biomarker panel validation
- Enzyme activity quantification
- Therapeutic protein monitoring across batches
With custom assay development and high dynamic range detection, this platform provides robust, reproducible measurements that integrate seamlessly with previous discovery datasets.
Unified QC and Bioinformatics Backbone
No matter which platform you choose, all services include:
- SOP-standardized sample processing
- Built-in quality checkpoints (CV, FDR, ID confidence)
- Customized data interpretation via GO, KEGG, Reactome
- Optional dashboard delivery and multi-omics overlays
💡 Not sure which platform fits your study?
Let our scientists match your biological question with the right technology. Whether you're profiling 5 samples or 500, we help you get from protein to insight—efficiently and accurately.
How to Prepare Samples for Proteomics Profiling
| Sample Type |
Minimum Input |
Recommended Conditions & Notes |
| Cultured Cells |
≥ 5 × 10⁶ cells |
Use fresh or snap-frozen pellets. For secretome analysis, culture in serum-free medium. |
| Animal or Plant Tissues |
≥ 50 mg |
Snap-frozen preferred. Avoid freeze-thaw cycles. Homogenization performed prior to protein extraction. |
| Organelle Fractions |
≥ 10 µg total protein |
Provide details on isolation method or request Creative Proteomics to perform fractionation. |
| Conditioned Media |
≥ 500 µL (serum-free) |
Suitable for secretome or exosome proteomics. Media should be free of serum proteins. |
| Exosome Pellets |
≥ 5 µg total protein |
Isolated using ultracentrifugation or kit-based methods. DNase/RNase pre-treatment optional. |
| Biofluids (e.g., plasma) |
≥ 50 µL |
Plasma/serum samples should be EDTA- or heparin-treated. For deep coverage, depletion may be advised. |
| FFPE Tissue Sections |
≥ 5 sections (10 µm thick) |
Must be freshly cut. Provide H&E reference if spatial localization is needed. |
Additional Notes:
- If you're unsure about sample quantity or buffer compatibility, our team can help optimize your protocol.
- For low-input samples (e.g., <5 µg protein), ultra-sensitive workflows are available—please contact us in advance.
- We support both standard protein extraction and subcellular/organelle purification as part of our full-service options.
What You'll Receive
- Raw MS data and peptide/protein identification files
- Quantitative protein matrix with normalized abundance values
- Subcellular localization annotation
- Pathway and GO enrichment analysis
- Protein-protein interaction and clustering reports
- Optional: Interactive dashboards or integrative omics overlays
All deliverables are tailored to your experimental design and output preferences, ensuring clarity, traceability, and scientific usability.

Representative demo results from subcellular proteomics analysis, including differential expression matrix, subcellular localization profiling, phosphorylation motif enrichment, and pathway-level biological interpretation.
Real-World Case Highlights
Case 1: Diurnal Nuclear Proteome Remodeling in Mouse Liver
- Objective: Investigate how circadian rhythm regulates nuclear protein expression and phosphorylation dynamics in liver tissue.
- Method: Nuclear protein isolation + SILAC-based quantitative LC–MS/MS + pathway enrichment analysis.
- Result: The study identified extensive rhythmic changes in nuclear protein abundance and phosphorylation, linking diurnal regulation to transcription, metabolism, and chromatin remodeling.
Read full article
Case 2: Hypoxia-Driven Remodeling of the Mitochondrial Proteome
- Objective: Examine how hypoxic stress alters the expression of mitochondrial proteins involved in translation and translocation in cancer cells.
- Method: Hypoxia treatment of HeLa cells + SILAC-based mitochondrial fraction proteomics + LC–MS/MS + GO annotation.
- Result: Hypoxia downregulated mitochondrial ribosomal proteins and translocases, indicating translational inhibition and mitochondrial remodeling under oxygen deprivation.
Read full article
FAQs – You May Want to Know
How do you ensure organelle purity during subcellular proteomics?
We use optimized differential centrifugation, gradient ultracentrifugation, and affinity purification combined with western blot validation of organelle-specific markers. Cross-contamination is evaluated using quantitative enrichment scoring and GO-based localization analysis.
Can you distinguish between cytosolic contaminants and truly secreted proteins in secretome studies?
Yes. We employ serum-free culture, EV depletion, and use tools like SignalP, SecretomeP, and WoLF PSORT for secretion pathway annotation. Additionally, contamination is flagged via intracellular marker abundance and excluded during downstream analysis.
How do you deal with batch effects in large-scale TMT or label-free projects?
We embed internal pooled references and run bridge channels for TMT-based studies. For label-free experiments, we use spike-in standards and global normalization strategies. Downstream correction includes ComBat and variance-stabilizing transformations to remove technical drift.
Can you identify and annotate isoforms or proteoforms?
Yes. When appropriate sequence databases are available, we support isoform-level identification using top-down or bottom-up integration pipelines. We also offer proteoform inference via PTM co-localization and alternative splicing patterns.
How do you handle uncharacterized or novel proteins in non-model species?
For non-model organisms or custom genomes, we support proteogenomics workflows using custom FASTA databases derived from RNA-seq or genome annotation. Novel peptides can be mapped to reference scaffolds and functionally annotated via orthology.
Can you help prioritize biomarker candidates from large datasets?
Yes. We use machine learning–based scoring (e.g., random forest, LASSO, SVM) to rank protein candidates based on expression stability, condition specificity, and functional relevance. This helps streamline validation and clinical translation.
What strategies do you use to increase proteome depth in challenging samples?
We combine multi-dimensional fractionation (Hi-pH reversed-phase, SCX), low-flow nanoLC, and longer gradients to improve peptide separation. For extremely complex matrices, we also support OFFGEL fractionation or organelle pre-enrichment.
Is your platform compatible with single-cell proteomics strategies?
While we do not offer true single-cell proteomics at this stage, we support ultra-low input workflows down to 1 µg total protein, which is suitable for sorted cell populations or rare tissue regions. Please contact us for project-specific feasibility.
Optional Bioinformatics & Interpretation Modules
| Bioinformatics Data Analysis Components |
Description |
| Quantitative Proteomics Analysis |
- Protein Abundance: Quantify protein levels within subcellular compartments, enabling relative expression comparisons.
- Differential Expression: Identify significantly altered proteins using statistical methods, aiding in biomarker or target discovery.
- Hierarchical Clustering: Organize proteins into clusters with similar expression patterns to unveil functional groups. |
| Post-translational Modification (PTM) Analysis |
- PTM Identification: Identify and characterize various PTMs (e.g., phosphorylation, glycosylation) to unravel regulatory mechanisms.
- Site Localization: Pinpoint the exact amino acid residues modified by PTMs for precise functional insights. |
| Functional Analysis |
- Gene Ontology (GO) Enrichment: Categorize proteins based on biological processes, molecular functions, and cellular components, revealing functional roles.
- Functional Annotation: Annotate proteins with known functions to explore their involvement in specific cellular processes or diseases.
- Pathway Enrichment: Identify enriched biological pathways among subcellular proteins to understand functional context. |
| Interaction Analysis |
- Protein-Protein Interaction (PPI) Analysis: Build protein-protein interaction networks to uncover key players and pathways within subcellular structures. |
| Subcellular Localization Prediction |
- Subcellular Localization Prediction: Predict the specific subcellular compartments where proteins are likely to reside, enhancing functional understanding. |
| Differential Gene Expression Analysis |
- Identification of Differentially Expressed Genes: Analyze gene expression data to identify genes showing significant expression changes within subcellular compartments. |
| Biomarker Discovery |
- Identification of Potential Biomarkers: Explore the dataset for proteins that may serve as potential biomarkers for diseases or biological processes. |
| Multi-Omics Integration |
- Integration with Other Omics Data: Combine subcellular proteomics data with other omics data types, such as genomics or metabolomics, for a comprehensive systems biology approach. |
| Subcellular Protein Distribution Visualization |
- Data Visualization: Utilize visualization tools to create subcellular protein distribution maps, enhancing the understanding of protein spatial localization. |
From System to Subcellular: Your Proteomics Partner
At Creative Proteomics, we bring together cutting-edge instrumentation, proprietary workflows, and experienced bioinformatics teams to offer not just protein lists, but functional, localized, and integrated protein insights. Whether you aim to track stress-induced proteome changes, decode cell signaling, or discover novel subcellular biomarkers, we deliver the resolution and reliability your research demands.
→ Get in touch now to customize your proteomics project. Let's turn your proteins into discoveries.