Title: BioID-based proteomic analysis of the Bid interactome identifies novel proteins involved in cell-cycle-dependent apoptotic priming
Journal: Cell Death & Disease
Published: 2020, Volume 11
Background
Apoptotic priming refers to how close a cell is to mitochondrial outer-membrane permeabilisation (MOMP), which controls whether cells undergo apoptosis. Cells in mitosis are primed for apoptosis, and this priming can be influenced by Bcl-2 family proteins. Specifically, the Bcl-2 family protein Bid becomes phosphorylated in mitosis, which increases apoptotic priming and sensitivity to antimitotic drugs. However, the regulation of apoptotic priming during mitosis, especially how Bid interacts with other proteins, is not well understood. This study uses a BioID proteomic screen to identify novel proteins involved in the regulation of apoptotic priming during mitosis, with a focus on Bid. The screen reveals that the outer mitochondrial protein VDAC2 plays a crucial role in regulating changes in apoptotic priming following Bid phosphorylation.
Materials & Methods
Cell Culture
Cell lines (MCF-7, BT-474, MDA-MB-231, SK-BR-3, MCF-10A, HEK-293T, HeLa).
MCF-10A cells were cultured in DMEM-F12 supplemented with specific growth factors and serum.
HeLa cells were grown in EMEM with 10% FBS, while other cell lines were cultured in DMEM with 10% FBS.
Single-Cell Fate Tracking
Cells were treated with Taxol, ABT737, or DMSO and imaged at 15-min intervals for 48–60 h using a Leica live-cell imaging system.
Mitosis, apoptosis, and slippage were identified based on cellular morphology using ImageJ.
Antibodies, Immunofluorescence, and Immunoblotting
Specific antibodies (e.g., anti-Bid, anti-Myc, anti-V5, anti-GFP) were used for immunoblotting and immunofluorescence.
Immunofluorescence was performed using AlexaFluor-conjugated secondary antibodies and visualized using wide-field or confocal microscopy.
Immunoblotting was performed using SDS-PAGE, and proteins were detected using LiCor Odyssey CLx imager.
Mitotic Enrichment
Cells were synchronized in mitosis using nocodazole (200 ng/mL for 16 h) and harvested by mechanical dislodgement.
Flow Cytometry for Cell Cycle Analysis
Cells were fixed, stained with DAPI, and analyzed using a BD Fortessa flow cytometer.
Data were analyzed using Modfit LT to determine cell cycle distribution.
BioID-Mediated Proximity Labelling
Cells expressing BirA*-fusion proteins were labeled with 50 µM biotin for 16 h.
For mass spectrometry, cells were lysed, and biotinylated proteins were purified using streptavidin beads.
Streptavidin-Affinity Purification
Biotinylated proteins were isolated using MagReSyn streptavidin beads, washed, and eluted for downstream analysis.
Mass Spectrometry and Protein Identification
Purified proteins were digested with trypsin and analyzed by LC-MS/MS using a Thermo Orbitrap Elite.
Data were processed using Progenesis QI and searched against SwissProt and Trembl databases.
Label-Free Protein Quantification and Data Analysis
Relative protein quantification was performed using Progenesis QI, and statistical analysis was conducted using ANOVA.
STRING Analysis
Predicted interaction networks for Bid were generated using the STRING database (v11.0).
VDAC2 Deletion by CRISPR/Cas9
VDAC2 was knocked out in MCF-7 cells using CRISPR/Cas9 with specific sgRNAs.
Clonal populations were screened for indels by PCR and sequencing.
Statistical Analysis
Data were analyzed using GraphPad Prism v7, with specific tests and sample sizes detailed in figure legends.
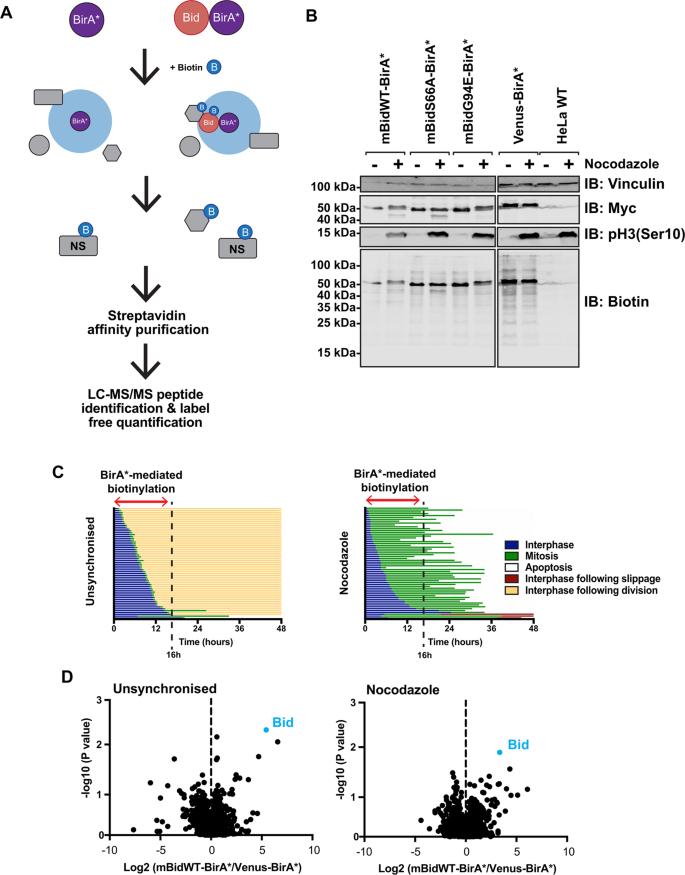 Schematic diagram of BioID labelling strategy.
Schematic diagram of BioID labelling strategy.
Results
BioID for Interrogating Bid Interactome in Live Cells
BioID (biotin-labelling identification) was employed to study Bid interactions in live cells, focusing on mitosis-specific regulation of apoptotic priming.
Bid phosphorylation (serine 66 in mouse, 67 in human) during mitosis increases apoptotic priming in colon carcinoma cells.
mBid–BirA* fusion proteins were generated to identify proximal interacting partners in live cells.
Validation of BioID Approach
Truncated Bid (tBid)–BirA* fusions retained pro-apoptotic activity and successfully biotinylated known binding partners (e.g., GFP-Bcl-XL).
Labeling specificity was confirmed using immunofluorescence and streptavidin-affinity purification.
Identification of Mitosis-Specific Bid Interactions
Quantitative mass spectrometry identified >400 proteins in each sample, with VDAC2 emerging as a key candidate enriched in mitotic cells expressing mBidWT–BirA*.
VDAC2 enrichment was significantly reduced in cells expressing non-phosphorylatable (mBidS66A) or BH3-domain-deficient (mBidG94E) Bid variants.
VDAC2 as a Key Bid Partner in Mitosis
VDAC2 co-localized with Bid in mitochondria, suggesting a functional interaction.
CRISPR/Cas9 knockout of VDAC2 in MCF-7 cells shifted cell fate from apoptosis to slippage during Taxol-induced mitotic arrest, mimicking the effect of Bid knockdown.
Restoration of VDAC2 expression in knockout cells rescued apoptotic priming.
Bid Phosphorylation and Apoptotic Priming
Bid phosphorylation was required for increased apoptotic priming in mitosis.
Knockdown of endogenous Bid (hBid) reduced apoptosis, which was rescued by expressing wild-type Bid (mBidWT-GFP) but not phosphorylation-deficient (mBidS66A-GFP) or BH3-domain-deficient (mBidG94E-GFP) variants.
BH3 mimetic ABT737 restored apoptosis levels in Bid knockdown and VDAC2 knockout cells, indicating Bid phosphorylation sets mitochondrial priming.
Functional Role of VDAC2 in Coupling Bid Phosphorylation to Apoptosis
VDAC2 knockout cells showed reduced Bak expression, which was restored upon VDAC2 re-expression.
VDAC2 deletion uncoupled Bid phosphorylation from apoptotic priming, as cells expressing mBidWT-GFP in a VDAC2 knockout background exhibited reduced apoptosis.
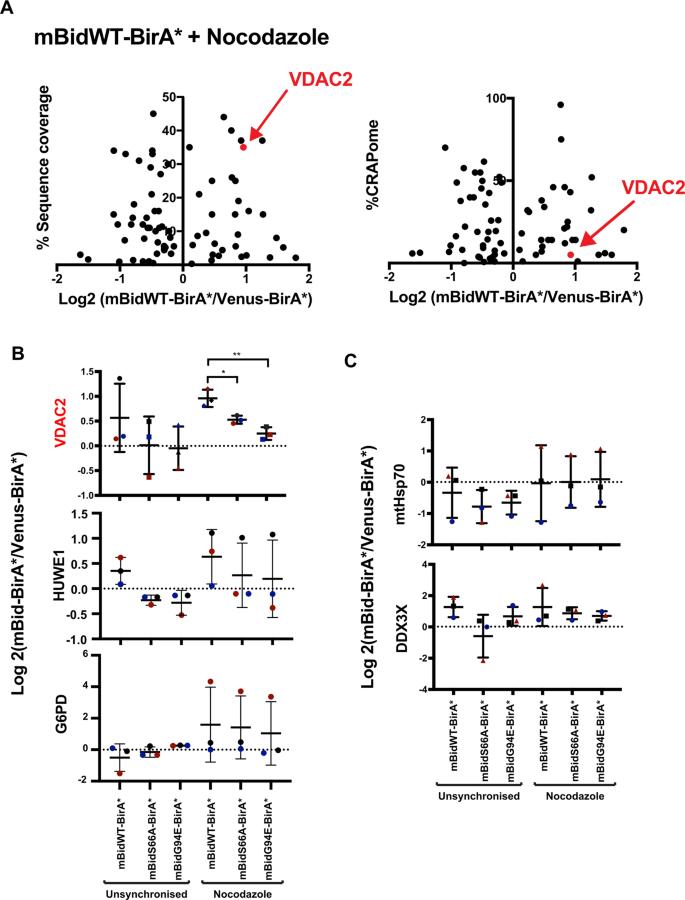 BioID identifies the voltage-dependent anion channel 2, VDAC2, as showing Bid phosphorylation-dependent enrichment in mitotic cells.
BioID identifies the voltage-dependent anion channel 2, VDAC2, as showing Bid phosphorylation-dependent enrichment in mitotic cells.
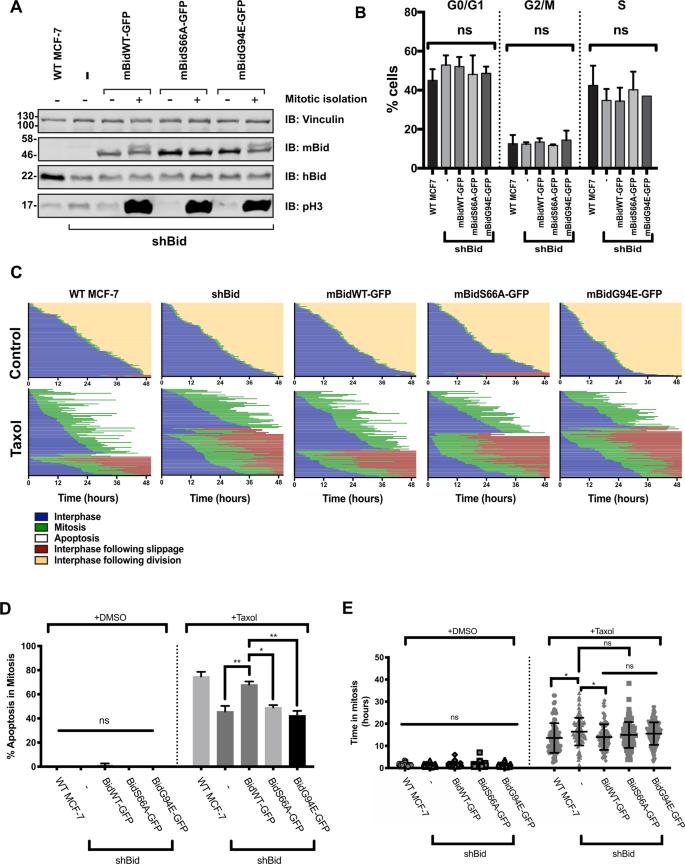 Phosphorylation of Bid regulates apoptotic priming in mitotic breast-cancer cells.
Phosphorylation of Bid regulates apoptotic priming in mitotic breast-cancer cells.
Reference
- Pedley, Robert, et al. "BioID-based proteomic analysis of the Bid interactome identifies novel proteins involved in cell-cycle-dependent apoptotic priming." Cell death & disease 11.10 (2020): 872.




