De Novo Antibody Sequencing Service Using High-Resolution LC-MS/MS
At Creative Proteomics, we deliver fast, accurate de novo antibody sequencing services powered by advanced LC-MS/MS platforms and proprietary data analysis workflows. Our platform is built for flexibility—tailored to meet the sequencing challenges of research teams in antibody discovery, biosimilar development, and protein engineering.
Unlike conventional approaches that rely on prior genetic information, de novo sequencing determines the complete amino acid sequence of an antibody directly from the protein itself. This method also enables the identification of post-translational modifications (PTMs)—such as glycosylation or oxidation—without requiring cDNA, hybridoma access, or reference databases.
Whether you're working with a legacy antibody lacking sequence documentation or need to verify critical variable regions for therapeutic design, our method provides high-confidence, research-grade sequences that support downstream cloning, expression, and optimization.
Submit Your Request Now
×
What You Get:
- Full heavy & light chain sequences—CDR-verified
- High coverage with PTM annotation
- Works with purified antibodies or serum
- Delivered fast. Designed for recombinant expression.
- What is
- Service Details
- FAQ
- Case Study
- Demo
- Publication
Unlocking Antibody Insights with De Novo Sequencing
In today's fast-moving landscape of therapeutic antibody discovery and protein engineering, researchers are increasingly faced with a critical challenge: how to recover or confirm the precise amino acid sequence of an antibody when no genetic template is available. Whether due to hybridoma loss, proprietary constraints, or incomplete sequence records, the need to reconstruct antibody sequences from scratch—de novo—has become a pivotal step in many biopharma and CRO workflows.
De novo antibody sequencing uses advanced mass spectrometry (MS)-based techniques to directly read peptide fragments from purified antibodies. This bypasses the need for DNA or mRNA templates, enabling scientists to recover heavy and light chain variable regions, especially complementarity-determining regions (CDRs), with high accuracy. As patent expirations, biosimilar development, and antibody repurposing expand across the industry, this technology is increasingly vital for:
- Generating recombinant antibody sequences from legacy or unknown sources
- Supporting antibody humanization and optimization strategies
- Ensuring functional integrity during antibody reformatting and QC
- Enabling regulatory documentation or IP filings for novel biologics
From global pharma teams working on next-generation biologics to academic labs salvaging precious hybridoma lines, de novo sequencing empowers scientists to regain control over their antibody assets—accurately, reproducibly, and without genetic constraints.
What Is De Novo Antibody Sequencing?
De novo antibody sequencing refers to the process of determining an antibody's amino acid sequence directly from the protein itself—without relying on existing genetic or sequence information. Unlike template-based sequencing, which aligns peptide fragments against known reference databases, de novo sequencing builds the sequence from scratch, using only the experimental data obtained from tandem mass spectrometry (MS/MS).
In protein research, "de novo" literally means "from the beginning." This method is particularly critical when:
- The original hybridoma cell line is lost, degraded, or contaminated
- No cDNA or mRNA is available for traditional sequencing
- The antibody originates from proprietary, non-disclosed sources
- Accurate sequence confirmation is required for biosimilar development or intellectual property filing
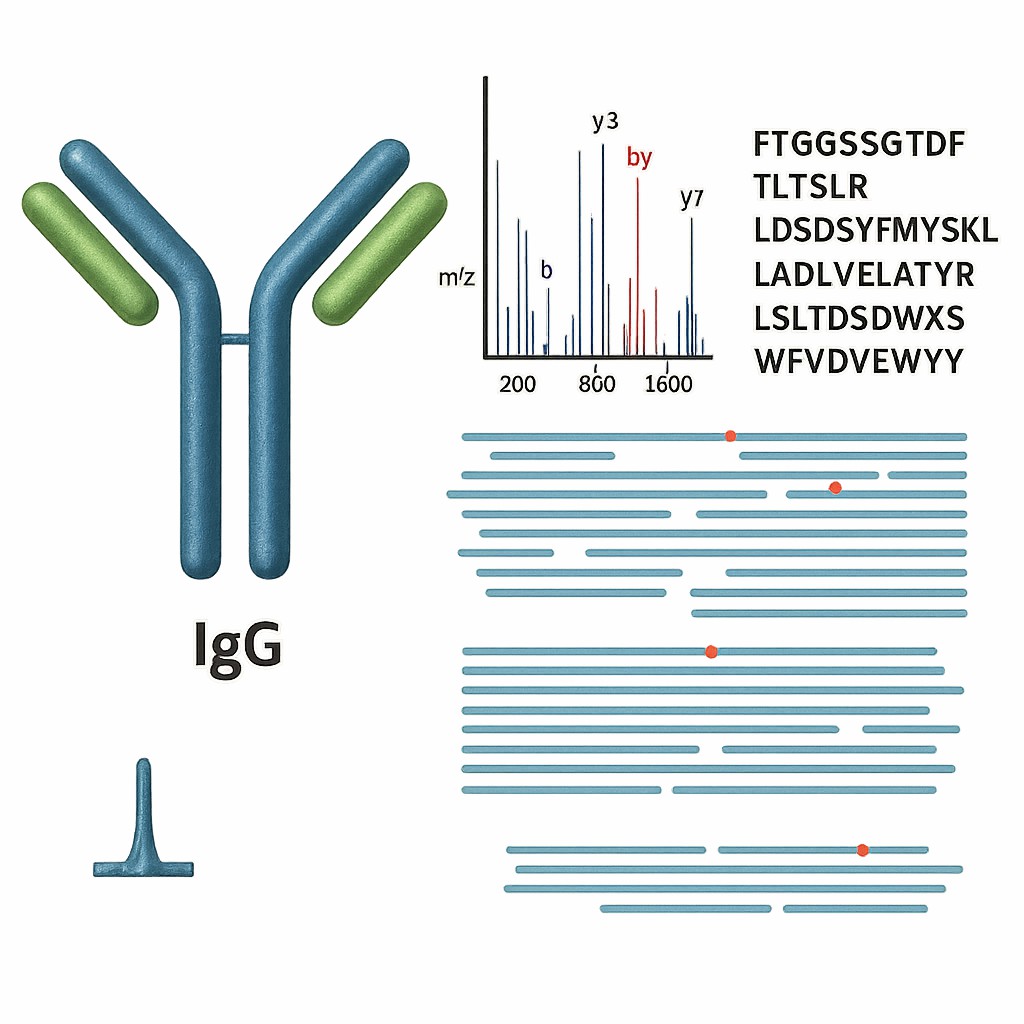
For antibodies, de novo sequencing must resolve not only the overall chain composition, but also the hypervariable CDR regions, which define antigen-binding specificity. This requires high-coverage MS data, careful peptide fragmentation (often via multiple digestion enzymes), and sophisticated software algorithms that can reconstruct overlapping peptide maps to assemble the full heavy (VH) and light (VL) chain sequences.
The end result is a template-independent, research-grade antibody sequence that enables recombinant expression, epitope mapping, functional studies, and more. It's a powerful rescue and validation tool for anyone working with unknown or legacy antibodies.
Our De Novo Antibody Sequencing Service
At Creative Proteomics, we offer flexible and accurate de novo antibody sequencing with minimal sample requirements. In most cases, just a few hundred micrograms of purified antibody—≥ 95% purity—is sufficient for full heavy and light chain sequence determination.
Our platform integrates multi-enzyme digestion strategies with high-resolution LC-MS/MS and proprietary databases to confidently differentiate closely related amino acids such as leucine and isoleucine. These distinctions are made based on enzyme-specific cleavage patterns and established frequency data from our in-house reference sets.
Beyond primary sequence coverage, we also provide detailed characterization of post-translational modifications (PTMs)—including glycosylation, oxidation, and other user-defined modifications. These insights are critical for antibody functional studies, expression stability assessments, or biosimilar comparison projects.
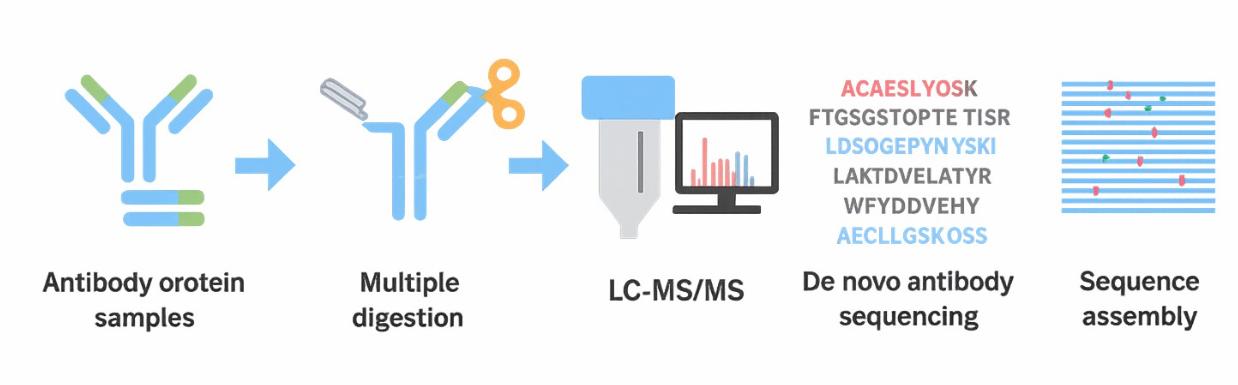
How Does De Novo Antibody Sequencing Work?
1. Sample Preparation
The process begins with a purified antibody sample, typically in the form of intact IgG or Fab fragments. To enhance sequence coverage, the antibody is digested using multiple complementary proteases (e.g., trypsin, chymotrypsin, Lys-C) to generate overlapping peptides that span the full sequence.
2. Peptide Fragmentation via Tandem MS
Each resulting peptide is subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS). Inside the instrument, peptides are ionized and fragmented using methods such as:
- Collision-Induced Dissociation (CID)
- Higher-energy Collisional Dissociation (HCD)
- Electron Transfer Dissociation (ETD)
These fragmentation strategies break peptides at different backbone locations, creating b- and y-ion series that reveal the amino acid sequence.
3. De Novo Peptide Assembly
Using sophisticated algorithms, de novo sequencing software analyzes MS/MS spectra to predict short peptide sequences (known as "tags") directly from the fragmentation data. These tags are then aligned and stitched together based on overlapping regions, enzyme specificity, and mass constraints.
4. Full-Length Sequence Reconstruction and Validation
The assembled peptide maps are used to reconstruct the complete heavy and light chain sequences, including variable (V), diversity (D), joining (J), and constant (C) regions where applicable. Manual curation is often employed to confirm critical regions like CDRs and to resolve ambiguous residues such as isoleucine/leucine.
When needed, additional fragmentation methods or orthogonal validation techniques may be used to ensure high sequence confidence.
Applications in Biopharma and CROs
✅ Recombinant Antibody Production from Lost or Legacy Sources
One of the most common use cases is recreating antibodies from hybridomas that are no longer viable or have lost productivity due to genetic drift or contamination. De novo sequencing enables the accurate reconstruction of heavy and light chain sequences, which can then be cloned and expressed recombinantly—ensuring consistent performance across batches.
✅ Biosimilar Development and Intellectual Property Strategy
Pharmaceutical companies developing biosimilars often require precise characterization of originator antibodies—especially when sequence data is not publicly disclosed. De novo sequencing provides high-confidence results that support:
- Competitive product profiling
- Patent circumvention or IP documentation
- Regulatory submissions requiring structural comparability
✅ Epitope Mapping and Antibody Humanization
When working with therapeutic candidates derived from animals, researchers often use de novo sequencing to guide CDR grafting and humanization efforts. Knowing the full sequence allows rational design of mutations to reduce immunogenicity while preserving target binding.
✅ Contract Research and QC for Outsourced Projects
CROs offering antibody production or engineering services may need to validate client-delivered antibodies for sequence fidelity, especially when working with legacy reagents or partially annotated sequences. De novo sequencing provides a definitive reference, enabling quality assurance, batch reproducibility, and traceability.
Whether you're recovering a legacy clone or de-risking biosimilar development, de novo sequencing ensures you know exactly what you're working with—amino acid by amino acid.
Service Features: Why Our De Novo Sequencing Delivers
Ultra-High Accuracy
- Distinguishes difficult amino acid pairs (e.g., Leu vs. Ile, Gln vs. Lys)
- Quantifies post-translational modifications and variant forms
Comprehensive Sequence Coverage
- Uses up to 8 proteases to ensure full antibody coverage
- 10× peptide reads for each CDR site to guarantee precision
Compatible with All Antibody Types
- Works with IgG, IgM, and non-standard formats
- Accurate sequencing of labeled antibodies (e.g., FITC, Biotin, GST-tagged)
Reliable for Any Species or Isoform
- Supports antibodies from human, mouse, rabbit, rat, camelid, and more
- Handles different antibody isoforms and subclasses with ease
Rigorous Quality Control
- Manual verification of key residues
- Strict validation from digestion to sequence assembly
Sample Requirements and Submission Guidelines
Sample Formats Accepted
- Purified antibody: IgG, Fab, or scFv
- Supernatant: Hybridoma culture media
- Serum/plasma-derived antibodies
- Labeled antibodies (e.g., FITC, Biotin, GST-tagged)
Minimum Quantity & Purity
| Sample Type | Minimum Amount | Recommended Purity |
|---|---|---|
| Purified antibody or Fab | ≥ 100 µg | ≥ 90% (SDS-PAGE) |
| Hybridoma supernatant | ≥ 500 µL | Cellular debris removed |
| Serum/plasma-derived IgG | ≥ 500 µL | Enrichment or depletion |
Use concentration methods (e.g., Protein A/G) for dilute samples.
Handling, Storage & Shipping
- Immediately freeze samples in liquid nitrogen post-preparation; store at –80 °C.
- Ship on dry ice to maintain low temperatures and avoid freeze–thaw cycles.
- Clearly label each tube with sample ID, concentration, buffer, date.
✅ Tips to Ensure Quality
- Use one species or sample per tube to prevent cross-contamination.
- For antibodies expressed in non-native systems: provide expression host details.
- Ensure supernatant/supplement buffer components are declared (e.g., serum proteins).
- For multi-protease digestion: we handle the process—no on-sample digestion needed beforehand.
Deliverables: What You'll Receive from Our Service
Full-Length Heavy & Light Chain Sequences
- Complete amino acid sequences in FASTA format (VH and VL variable regions plus constant regions)
- Both heavy and light chain alignments, ready for cloning or expression
- Confirmed sequence coverage supporting research-grade applications
Sequence Coverage Maps & Confidence Scores
- Interactive peptide-by-peptide mapping showing depth of coverage (>10× in CDRs and >5× elsewhere)
- Confidence scores for each peptide and residue, including flagged ambiguities
- Data visualization to facilitate in silico engineering and quality validation
Post-Translational Modification & Label Annotations
- Identification of relevant PTMs (e.g., glycosylation, oxidation, deamidation)
- Reporting of any small labels (e.g., FITC, Biotin) or fusion tags (e.g., GST) incorporated in the sample
Raw MS/MS Data (Optional)
- Upon request, deliverables can include all raw LC–MS/MS spectra and result files
- Enables integration into client pipelines, additional analysis, or reprocessing
Recombinant Antibody Expression (Optional Add-On)
- Optionally, we can perform expression and purification of the identified sequence
- Receive recombinant antibody protein—fully validated and ready for downstream use
FAQs: De Novo Antibody Sequencing Clarified
Can you sequence antibodies directly from serum, or do I need a monoclonal sample?
De Novo sequencing is most reliable with a single monoclonal antibody—mixed polyclonal samples can complicate data. For serum samples, targeted purification (e.g., antigen affinity pull-down) is recommended to isolate a specific antibody clone before sequencing.
How confident can I be in critical regions like CDRs?
Our process requires ≥ 10× overlapping peptide evidence for every CDR residue, ensuring that hypervariable regions—crucial for antigen binding—are accurately determined.
Will labeled or modified antibodies interfere with sequencing?
No. Our MS workflows and software pipelines are optimized to recognize and account for small tags (e.g., FITC, Biotin) or large fusion partners (e.g., GST), enabling precise core-sequence determination even in labeled samples .
How do you resolve amino acid isomers like Leu/Ile or Gln/Lys?
Thanks to high-resolution MS/MS and advanced fragmentation schemes (e.g., HCD + ETD), we can distinguish leucine from isoleucine and glutamine from lysine, overcoming limitations of low-resolution platforms.
My antibody has unusual PTMs—can you still sequence it?
Yes. We routinely detect and map post-translational modifications such as glycosylation, oxidation, and deamidation. These modifications are incorporated into peptide assembly and reported with confidence scores.
What if a residue remains ambiguous after MS analysis?
Ambiguous assignments trigger manual expert review and may prompt additional fragmentation or enzyme digest to resolve uncertainty. Our QC ensures that no questionable residue is left unresolved.
Learn about other Q&A.
Case Study: De Novo Sequencing Identifies Functional Antifungal Protein Sin a 1 from Mustard Seeds

Client Institution: Munster Technological University & University College Cork
Publication: Biochemistry and Biophysics Reports, 2022
DOI: 10.1016/j.bbrep.2022.101208
- Background
- Methods
- Key Results
- Conclusion
A research team aimed to isolate and functionally characterize natural antifungal proteins from white mustard (Brassica hirta) seeds. After a series of protein purification steps, they obtained a ~14 kDa protein with strong anti-yeast activity. However, the amino acid sequence of this protein was unknown, and no reference genome or cDNA data were available for direct gene-based identification.
To proceed with sequence validation and isoform comparison, the team required accurate, full-length de novo protein sequencing from the purified sample.
Creative Proteomics' Contribution
To support this discovery, Creative Proteomics performed full-service de novo sequencing of the purified antifungal protein:
- In-gel digestion of SDS-PAGE bands using six complementary proteases (Trypsin, Chymotrypsin, Glu-C, Arg-C, Lys-N, Pepsin)
- High-resolution nano LC-MS/MS analysis of digested peptide fragments
- De novo sequence assembly and analysis using PEAKS Studio v8.5 software
- Final sequence output in FASTA format, suitable for BLASTp homology searches and multiple sequence alignments
This approach provided the client with a complete and confident amino acid sequence, enabling functional annotation and classification.
Protein Extraction: Ammonium sulfate precipitation, thermal denaturation, and cation-exchange chromatography on AKTA system
Purity Check: SDS-PAGE under reducing and non-reducing conditions revealed a 14 kDa protein consisting of two disulfide-linked chains
De Novo Sequencing: Outsourced to Creative Proteomics—full-length sequence obtained (see Figure 3 of publication)
Bioinformatics: BLASTp search identified 96% identity with Sin a 1.0106, a 2S albumin family member
Antimicrobial Testing: MICs determined against yeasts, moulds, and bacteria
Mechanistic Assays: Yeast colony count, membrane permeability (PI staining), nucleotide leakage
Safety & Stability Tests: Caco-2 cytotoxicity, red blood cell hemolysis, resistance to proteases, heat, pH, and salt stress
| Key Finding | Details |
|---|---|
| Sequence Identity | 96% match to Allergen Sin a 1.0106 from Sinapis alba |
| Protein Identity | Confirmed as a Napin family isoform with antifungal properties |
| Spectrum of Activity | Active against Z. bailii, S. cerevisiae, F. culmorum; no antibacterial effect |
| Mechanism of Action | Membrane permeabilization and nucleotide leakage confirmed |
| Biostability | Retains antifungal activity after heat (100°C), pH shifts, and digestion |
| Safety | Non-toxic to human intestinal cells and red blood cells |
 Figure: Full-length amino acid sequence of the Sin a 1 protein as identified by Creative Proteomics' de novo LC-MS/MS sequencing. This enabled downstream homology analysis and functional validation.
Figure: Full-length amino acid sequence of the Sin a 1 protein as identified by Creative Proteomics' de novo LC-MS/MS sequencing. This enabled downstream homology analysis and functional validation.
Thanks to the de novo protein sequencing platform provided by Creative Proteomics, the research team:
- Successfully identified the protein as a functional isoform of Allergen Sin a 1
- Published the report of antifungal activity for this protein in a peer-reviewed journal
- Confirmed that plant-derived storage proteins can serve as promising antifungal agents
- Gained a high-purity, heat-stable, and non-toxic candidate with potential applications in agriculture or food biopreservation
Demo

Publication
Here are some publications in protein sequencing from our clients:

- Isolation of the mustard Napin protein Allergen Sin a 1 and characterisation of its antifungal activity. Mignone, G., Shwaiki, L. N., Arendt, E. K., & Coffey, A. Biochemistry and Biophysics Reports, 2022. https://doi.org/10.1016/j.bbrep.2022.101208
- Chemoproteomic identification of CO₂-dependent lysine carboxylation in proteins King, D. T., Zhu, S., Hardie, D. B., Serrano-Negrón, J. E., Madden, Z., Kolappan, S., & Vocadlo, D. J. Nature Chemical Biology, 2022 https://doi.org/10.1038/s41589-022-01043-1
- Identification of a novel anti-HIV-1 protein from Momordica balsamina leaf extract Coleman, M. I., Khan, M., Gbodossou, E., Diop, A., DeBarros, K., Duong, H., ... & Powell, M. D. International Journal of Environmental Research and Public Health, 2022. https://doi.org/10.3390/ijerph192215227