Plant Metabolomics Service
From stress adaptation to crop trait engineering, every research question demands precise metabolic insights. Creative Proteomics's advanced LC-MS/MS and GC-MS workflows transform raw complexity into actionable knowledge—fast, accurate, and publication-ready.
- Comprehensive Coverage – Primary metabolites, secondary pathways, phytohormones, and lipids in one platform
- Data You Can Trust – Orbitrap MS with<2 ppm mass accuracy; CV ≤10% for reproducible quantification
- Pathway-Centric Insights – Full integration with KEGG, PlantCyc, and advanced statistical modeling (PCA, PLS-DA)
- Custom-Fit Solutions – Targeted, untargeted, and pathway-focused options for every project scale
Submit Your Request Now
×- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- Case
- FAQ
What Is Plant Metabolomics and Why It Matters for Your Research
Plant metabolomics focuses on the comprehensive detection and quantification of small molecules in plants, covering primary metabolites like sugars, amino acids, and organic acids, as well as secondary metabolites such as flavonoids, terpenoids, and alkaloids. These metabolites are central to plant growth regulation, stress adaptation, and biosynthesis of valuable compounds, making metabolomics an indispensable approach for modern plant science.
Creative Proteomics delivers advanced LC-MS/MS and GC-MS–based plant metabolomics solutions that combine broad metabolite coverage with high sensitivity and reproducibility. Our service supports research in stress physiology, crop improvement, nutritional analysis, and metabolic pathway engineering, providing data-driven insights for both fundamental studies and applied agricultural innovation.
What Research Problems Does This Service Solve?
- Comparing genotypes or treatments to identify key metabolic differences
- Validating engineered pathways for targeted compound production
- Screening biomarkers for stress, nutrient efficiency, or disease resistance
- Improving product quality for food, pharma, or bio-based materials
Creative Proteomics Plant Metabolomics Solutions
- Untargeted Plant Metabolomics
- Targeted Plant Metabolomics
- Widely Targeted Metabolomics
- Secondary Metabolite Profiling
- Phytohormone Analysis
- Lipid & Fatty Acid Metabolomics
- Pathway-Focused Analysis
Comprehensive metabolite profiling without predefined targets, enabling discovery of novel compounds and global metabolic patterns.
Core Features:
- Detects thousands of metabolites across primary and secondary pathways
- Suitable for exploratory studies and hypothesis generation
Applications:
- Comparative analysis of genotypes or treatments
- Stress physiology and metabolic diversity research
Absolute quantification of selected metabolites using validated reference standards.
Core Features:
- High sensitivity and accuracy with internal standards
- Precise concentration measurement for defined metabolites
Applications:
- Nutritional quality assessment
- Validation of candidate biomarkers or engineered pathways
High-throughput identification and quantification of hundreds of metabolites in a single run.
Core Features:
- Combines broad coverage with quantitative accuracy
- Utilizes custom metabolite libraries and advanced triple quadrupole MS
Applications:
- Large-scale plant metabolome profiling
- Trait association and breeding research
Secondary Metabolite Profiling
In-depth characterization of specialized plant metabolites related to defense, pigmentation, and bioactivity.
Core Features:
- Wide coverage of phenylpropanoids, terpenoids, alkaloids, and more
- Advanced MS/MS spectral matching for confident identification
Applications:
- Functional ingredient validation
- Bioactive compound discovery in crops and medicinal plants
Phytohormone Analysis
Quantitative profiling of plant hormones and signaling molecules involved in growth and stress responses.
Core Features:
- High-sensitivity detection of low-abundance phytohormones
- Supports both developmental biology and stress adaptation studies
Applications:
- Hormonal regulation research
- Investigating signal transduction under abiotic or biotic stress
Lipid & Fatty Acid Metabolomics
Analysis of lipid composition and dynamics to understand energy storage and membrane function in plants.
Core Features:
- Comprehensive coverage of structural and signaling lipids
- Suitable for both polar and non-polar lipid classes
Applications:
- Stress adaptation studies involving membrane remodeling
- Oil crop quality evaluation and metabolic engineering
Pathway-Focused Analysis
Customized panels for rapid quantification within selected metabolic pathways.
Core Features:
- Targeted LC-MS/MS detection with high reproducibility
- Tailored to carbon metabolism, amino acid biosynthesis, or secondary pathways
Applications:
- Functional genomics validation
- Hypothesis-driven research projects
List of Detected Metabolites in Plant Metabolomics
| Category | Subclasses | Representative Compounds |
|---|---|---|
| Carbohydrates | Monosaccharides, Disaccharides, Sugar Alcohols | Glucose, Fructose, Sucrose, Maltose, Sorbitol, Mannitol |
| Organic Acids | TCA Cycle Intermediates, Dicarboxylic Acids | Citric Acid, Malic Acid, Succinic Acid, Fumaric Acid, Oxalic Acid |
| Amino Acids & Derivatives | Essential & Non-essential Amino Acids | Glutamic Acid, Aspartic Acid, Lysine, Proline, Methionine, Tryptophan |
| Nucleotides & Nucleosides | Purines, Pyrimidines, Phosphate Derivatives | Adenosine, Guanosine, Cytidine, Uridine, AMP, GMP |
| Polyamines | Diamines, Triamines | Putrescine, Spermidine, Spermine |
| Vitamins & Cofactors | Water- & Fat-Soluble Vitamins | Vitamin C, Vitamin E, Niacin, Riboflavin |
| Phenolic Compounds | Phenylpropanoids, Hydroxycinnamic Acids | Caffeic Acid, Ferulic Acid, p-Coumaric Acid, Chlorogenic Acid |
| Flavonoids | Flavonols, Flavones, Anthocyanins | Quercetin, Kaempferol, Luteolin, Cyanidin |
| Terpenoids | Monoterpenes, Diterpenes, Triterpenes | Geraniol, Gibberellins, Squalene |
| Alkaloids | Pyridine, Indole, Isoquinoline Alkaloids | Nicotine, Indole-3-acetic acid (IAA), Berberine |
| Phytohormones | Auxins, Cytokinins, Gibberellins, ABA, Jasmonates | Indole-3-acetic Acid (IAA), Zeatin, GA3, Abscisic Acid, Jasmonic Acid |
| Lipid Components | Fatty Acids, Glycerolipids, Phospholipids | Linoleic Acid, Oleic Acid, Phosphatidylcholine, Galactolipids |
| Sterols | Plant Sterols | β-Sitosterol, Stigmasterol |
| Specialized Metabolites | Glucosinolates, Isoflavones, Tannins | Sinigrin, Genistein, Catechin |
Why Choose Creative Proteomics for Plant Metabolomics Analysis?
- Broad Metabolite Coverage: Detect >1,000 metabolites spanning primary and secondary pathways.
- Ultra-High Sensitivity: Achieve LOD down to femtomole levels for low-abundance compounds using high-resolution MS.
- Exceptional Mass Accuracy: < 2 ppm error with Orbitrap MS and Q-TOF platforms ensures reliable metabolite identification.
- High Throughput Capacity: Process 300+ plant samples per batch, accelerating time to results for large-scale studies.
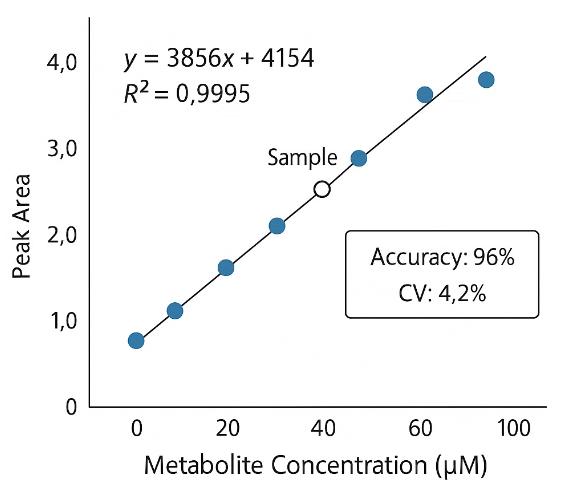
- Excellent Quantitative Reproducibility: Coefficient of Variation (CV) ≤10% across biological replicates.
- Comprehensive Statistical Analysis: PCA, PLS-DA, and pathway enrichment integrated into final reports.
Plant Metabolomics Analysis Workflow: Step-by-Step Process
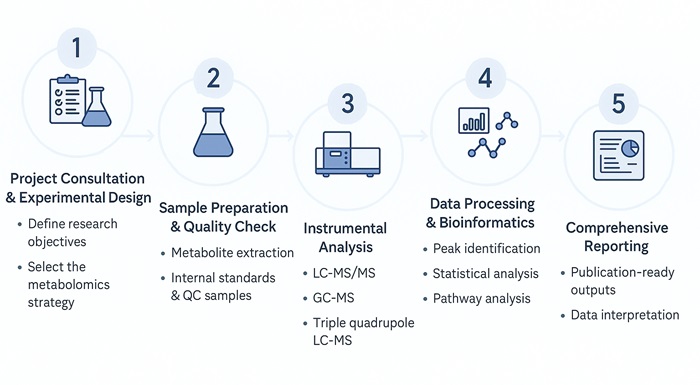
What Methods Are Used for Plant Metabolomics Analysis
LC-MS/MS for Semi-Polar and Polar Metabolites
- UHPLC System: Thermo Fisher Vanquish™
- Mass Spectrometer: Orbitrap Exploris™ 480 or Q Exactive™ HF-X
- Mass Range: m/z 70–1,200
- Resolution: 120,000 FWHM at m/z 200
- Polarity: Positive and negative ion switching
- Retention Time Precision: RSD ≤ 0.2%
- Quantification: External calibration with isotope-labeled internal standards
 Thermo Fisher Q Exactive (Figure from Thermo Fisher)
Thermo Fisher Q Exactive (Figure from Thermo Fisher)
GC-MS for Volatile and Derivatized Compounds
- GC System: Agilent 7890B
- MS Detector: Agilent 5977B Inert Plus EI/CI MSD
- Column: HP-5MS UI capillary (30 m × 0.25 mm × 0.25 μm)
- Ionization Mode: EI, 70 eV
- Mass Range: m/z 50–600
- LOD: < 1 ng/mL for most derivatized metabolites
 Agilent 7890B-5977B (Figure from Agilent)
Agilent 7890B-5977B (Figure from Agilent)
Triple Quadrupole LC-MS for Targeted Quantification
- System: AB Sciex QTRAP® 6500+
- Mode: MRM (Multiple Reaction Monitoring)
- LOD: Sub-picomole detection for plant hormones and signaling molecules
- Throughput: Up to 200 metabolites in one run
 SCIEX Triple Quad™ 6500+ (Figure from Sciex)
SCIEX Triple Quad™ 6500+ (Figure from Sciex)
Sample Requirements for Plant Metabolomics Service
| Parameter | Requirement |
|---|---|
| Accepted Sample Types | - Fresh or frozen plant tissues: leaves, stems, roots, seeds, fruits - Plant callus or cell cultures - Plant-derived fluids (sap, exudates) - Plant culture media |
| Recommended Quantity | - Tissue samples: ≥100 mg dry weight or ~500 mg fresh weight per sample - Fluids: ≥1 mL per sample |
| Replicates | Minimum 3 biological replicates per experimental group; technical replicates optional |
| Sample Collection | - Harvest samples rapidly to prevent metabolite degradation - Remove soil or debris immediately - Avoid cross-contamination between samples |
| Sample Pretreatment | - No chemical preservatives or stabilizers - For fresh tissues: freeze in liquid nitrogen immediately after collection |
| Homogenization | - Do not homogenize prior to shipping unless agreed upon - If homogenized, use precooled equipment and freeze-dry if possible |
| Storage Conditions | - Short-term: -20 °C (up to 24 h) - Long-term: -80 °C - Avoid freeze–thaw cycles |
| Transport Conditions | - Ship on dry ice in leak-proof, clearly labeled containers - Include an itemized sample list and experimental details |
| Additional Notes | - Provide information on treatment conditions (e.g., stress, chemical exposure) - Indicate if isotope labeling or special preprocessing was applied |
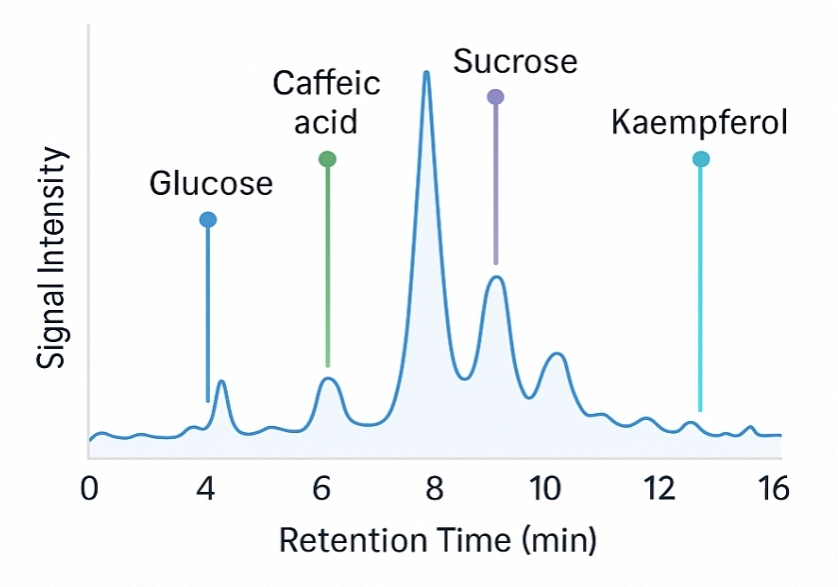
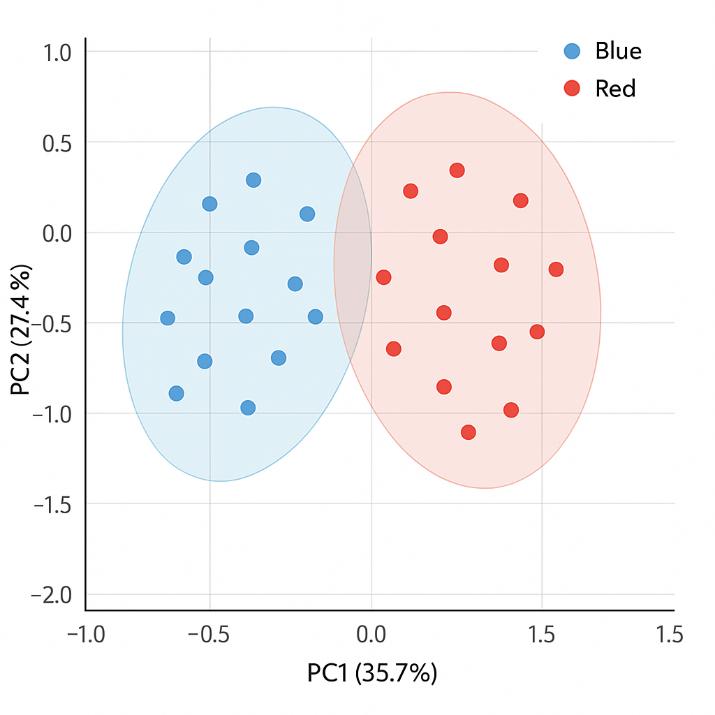
Demo Results
A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus.

Journal: Plant Cell
Published: 2011
- Background
- Methods
- Results
- Conclusion
- Reference
Drought stress remains one of the most significant threats to global agriculture. Understanding the biochemical regulation that enables certain plants to tolerate severe dehydration is critical for developing resilient crop systems. Sporobolus stapfianus (desiccation-tolerant, DT) and its closely related species Sporobolus pyramidalis (desiccation-sensitive, DS) provide an ideal sister-group contrast to dissect adaptive metabolic responses during dehydration.
Approach: Untargeted global metabolomic profiling was performed on young leaves at different hydration states:
- Fully hydrated (100% RWC)
- 60% RWC (early dehydration)
- Progressive drying stages to ~5% RWC for S. stapfianus
Analytical Platforms:
- UHPLC-MS/MS² (positive and negative ion modes)
- GC-MS for volatile and derivatized compounds
Metabolite annotation was achieved through MS/MS spectral matching against KEGG, PlantCyc, and proprietary databases. Statistical differentiation used PLS-DA (R² = 0.65, Q² = 0.61).
Metabolic Preconditioning in S. stapfianus
- Even under full hydration, S. stapfianus exhibited high baseline levels of amino acids and osmolytes (e.g., Asn, Gln) and lower energy metabolism intermediates compared to S. pyramidalis.
- Asparagine concentration was 451-fold higher than in the sensitive species, indicating nitrogen storage priming.
Early Dehydration Response (60% RWC)
- Rapid accumulation of proline (+5×), branched-chain amino acids, raffinose-family oligosaccharides, and sugar alcohols (arabitol, mannitol).
- β-Tocopherol, a lipophilic antioxidant, increased >3-fold, suggesting early oxidative stress defense activation.
- In contrast, S. pyramidalis showed minimal metabolic reprogramming at this stage.
Severe Dehydration (<40% RWC to Dry State)
- Glutathione and γ-glutamyl dipeptides skyrocketed (up to 1,873-fold) during advanced dehydration, indicating a robust antioxidant and nitrogen recycling response.
- Novel metabolite ophthalmate (linked to glutathione biosynthesis) was detected for the first time in plants.
- Lipid-soluble antioxidants surged: δ-tocopherol rose 89-fold, while lysolipids increased up to 45-fold, implying membrane remodeling.
- Sugars (sucrose, raffinose, stachyose) accumulated dramatically (10× to 74×), contributing to osmoprotection.
The untargeted metabolomics strategy revealed that desiccation tolerance in S. stapfianus is underpinned by:
- Metabolic priming with nitrogen-rich compounds
- Rapid induction of osmolytes and antioxidants during early dehydration
- Massive antioxidant surge and membrane stabilization under extreme drying
- Discovery of novel metabolites (ophthalmate), opening new research avenues in stress biology
Reference
- Oliver, Melvin J., et al. "A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus." The Plant Cell 23.4 (2011): 1231-1248.
FAQ of Plant Metabolomics Analysis Service
How do I decide between untargeted, targeted, and widely targeted metabolomics?
Untargeted metabolomics is ideal for discovery-driven projects where you need a broad overview of metabolic changes. Targeted analysis is best for hypothesis-driven studies requiring absolute quantification of specific metabolites. Widely targeted approaches combine both advantages for high-throughput quantitative profiling across hundreds of metabolites.
Can you analyze rare or species-specific secondary metabolites?
Yes. We maintain extensive in-house spectral libraries and leverage MS/MS validation to identify rare metabolites, even those with limited public database references. Custom libraries can also be developed for unique plant species.
What measures are taken to ensure reproducibility across biological replicates?
We apply rigorous quality control, including the use of isotope-labeled internal standards, pooled QC samples, and method calibration with CV ≤10% across replicates.
Can metabolomics results be integrated with transcriptomics or proteomics data?
Absolutely. We provide multi-omics integration and correlation analysis, enabling pathway-level interpretation and network visualization for systems biology studies.
What types of statistical analysis are included in your reports?
Reports typically include PCA, PLS-DA, hierarchical clustering, volcano plots, and pathway enrichment analysis. Advanced modeling or machine learning approaches can be requested as optional add-ons.
How do you confirm metabolite identification?
Metabolite annotation is performed using high-resolution MS, retention time alignment, and MS/MS spectral matching against KEGG, HMDB, PlantCyc, and our proprietary databases. Confidence scores and annotation levels are included in the report.
Do you support stable isotope labeling experiments for flux analysis?
Yes, we support ^13C or ^15N tracer-based metabolomics for metabolic flux studies. Please consult with us during project planning for isotope labeling protocols.
Can pathway-focused analysis be customized for specific research questions?
Yes. We offer fully customizable panels for specific biosynthetic pathways such as phenylpropanoid, flavonoid, or hormone signaling pathways, tailored to your experimental objectives.
What strategies are in place to handle matrix complexity in plant tissues?
We optimize extraction protocols for different plant matrices and apply advanced chromatographic separation to minimize matrix effects, ensuring accurate metabolite detection and quantification.
Learn about other Q&A about proteomics technology.
Publications
Here are some of the metabolomics-related papers published by our clients:

- Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy. 2023.
- A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B. 2023.
- The activity of the aryl hydrocarbon receptor in T cells tunes the gut microenvironment to sustain autoimmunity and neuroinflammation. 2023.
- Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. 2023.
- Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing. 2023.
- Anti-inflammatory activity of black soldier fly oil associated with modulation of TLR signaling: A metabolomic approach. 2023.
- Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium rhizosphaerae sp. 2023.