Glycerolipids Analysis Service
Complex lipid networks influence energy metabolism, membrane stability, and signaling in every living system. Inaccurate lipid profiling can compromise your research or product validation. Creative Proteomics provides high-resolution LC–MS/MS glycerolipid analysis with quantitative rigor, ensuring data you can trust for metabolic research, food stability assessment, and pathway interpretation.
Key Advantages:
- Comprehensive Coverage: Identify 1,500+ glycerolipid species in one run.
- Quantitative Precision: CV ≤ 15%, LOD down to 5 pg for trace detection.
- Advanced Technology: Orbitrap HF-X and SWATH® DIA for deep, reproducible coverage.
- Actionable Insights: Deliverables include annotated lipid lists, PCA plots, pathway maps.
Submit Your Request Now
×
Deliverables
- Raw Data Files: .raw / .wiff + mzML conversions.
- Processed Data Tables: Annotated glycerolipid species with quantitative values.
- Statistical Visuals: Heatmaps, PCA plots, volcano plots.
- Interpretive Report: Pathway mapping, class distribution charts, and QC summary.
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- FAQ
What Are Glycerolipids and Why Do They Matter?
Glycerolipids are esters formed by attaching fatty acids to a glycerol backbone, creating molecules that fulfill essential roles in biological systems:
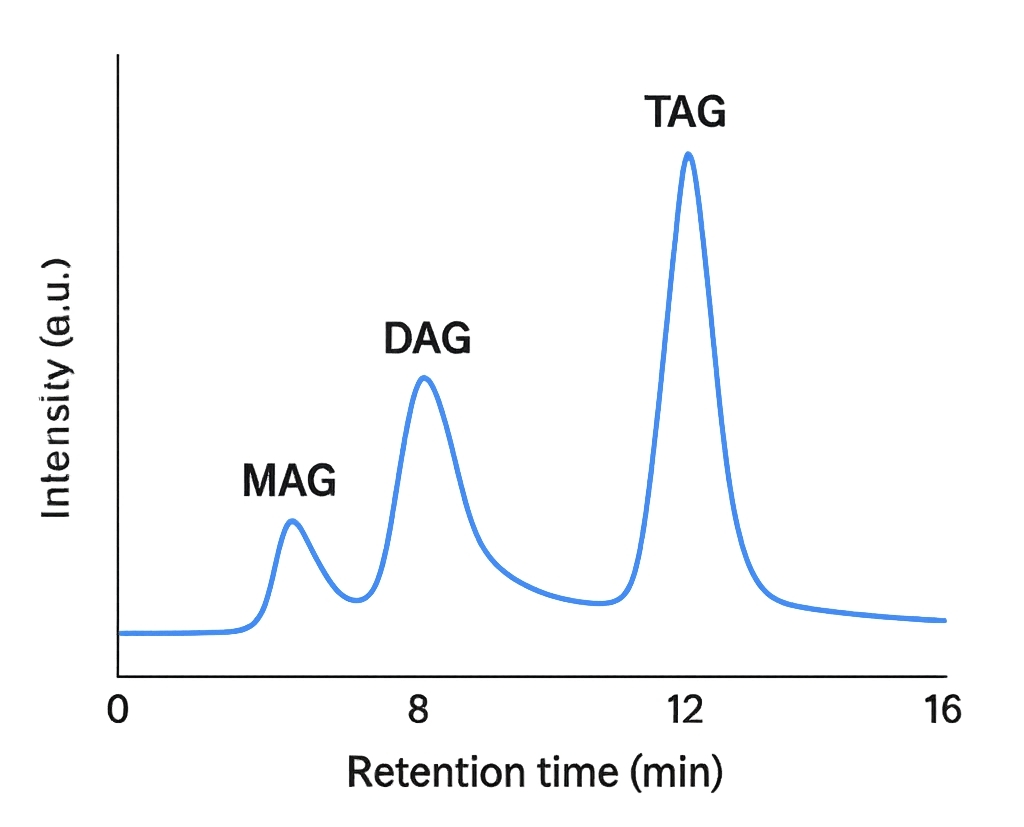
- Energy Storage: Triacylglycerols (TAGs) store metabolic energy in lipid droplets.
- Signal Regulation: Monoacylglycerols (MAGs) and diacylglycerols (DAGs) act as key messengers in lipid signaling networks.
- Membrane Integrity: Complex glycolipids such as MGDG, DGDG, and SQDG stabilize membranes and enable adaptive remodeling under stress.
Their structural diversity links energy metabolism, signaling, and cell structure. Accurate analysis of these molecules is critical for understanding metabolic pathways, validating functional food products, and exploring stress responses in plants or microbes.
Why Do You Need Glycerolipid Analysis?
Researchers and industrial clients use glycerolipid profiling to answer critical questions:
- Is my engineered organism efficiently channeling resources into TAG production for biofuel applications?
- Which signaling lipids fluctuate during stress or drug exposure?
- Can lipid degradation patterns confirm product stability in nutraceutical or food formulations?
- How do thylakoid lipids remodel under salinity, drought, or nitrogen starvation in plants?
Creative Proteomics provides data-driven insights that directly support these objectives.
Creative Proteomics's Glycerolipids Analysis Solutions
Untargeted Glycerolipid Profiling
Global lipidome coverage using high-resolution LC–MS/MS for qualitative and semi-quantitative assessment of thousands of glycerolipid species. Ideal for discovery-driven studies and comparative lipidomics.
Targeted Quantitative Analysis
Absolute quantification of major glycerolipid classes (TAG, DAG, MAG, and glycolipids) using isotope-labeled internal standards to ensure high precision and reproducibility.
Class-Specific Analysis
In-depth characterization of specialized lipid subclasses such as monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), sulfoquinovosyldiacylglycerol (SQDG), phosphatidylglycerol (PG), and DGTS.
Isomer and Structural Elucidation
Advanced MS/MS fragmentation and retention-time modeling to resolve isomeric species and confirm structural identities.
Comparative and Pathway Analysis
Differential profiling of glycerolipid changes across experimental conditions, integrated with KEGG and LipidMaps pathways for biological interpretation.
Customized Panels
Tailor-made analysis plans to focus on specific lipid subclasses, signaling intermediates, or metabolic markers relevant to your research objectives.
What Glycerolipids Can We Detect?
| Glycerolipid Class | Representative Subclasses | Example Species (Common Notation) |
|---|---|---|
| Triacylglycerols (TAG) | Neutral lipids storing energy | TAG 48:0, TAG 50:1, TAG 52:2, TAG 54:3, TAG 56:6 |
| Diacylglycerols (DAG) | Lipid signaling intermediates | DAG 32:0, DAG 34:1, DAG 36:2, DAG 38:4 |
| Monoacylglycerols (MAG) | Intermediates in lipid metabolism | MAG 16:0, MAG 18:1, MAG 18:2, MAG 20:4 |
| Monogalactosyldiacylglycerols (MGDG) | Chloroplast glycolipids for membrane structure | MGDG 34:3, MGDG 36:4, MGDG 38:6 |
| Digalactosyldiacylglycerols (DGDG) | Glycolipids in thylakoid membranes | DGDG 34:3, DGDG 36:4, DGDG 38:6 |
| Sulfoquinovosyldiacylglycerols (SQDG) | Sulfolipids important in photosynthesis | SQDG 32:0, SQDG 34:3, SQDG 36:6 |
| Phosphatidylglycerol (PG) | Structural phospholipids in chloroplasts | PG 32:0, PG 34:1, PG 36:4 |
| Diacylglyceryltrimethylhomoserine (DGTS) | Betaine lipids acting as membrane components | DGTS 32:0, DGTS 34:1, DGTS 36:4 |
| Lysoglycerolipids | Mono-substituted glycerol derivatives | Lyso-MGDG 18:3, Lyso-DGDG 18:3 |
| Acylated Galactolipids | Modified glycolipids for stress adaptation | Acyl-MGDG 54:8, Acyl-DGDG 56:9 |
Advantages of Our Glycerolipids Analysis Services
- Deep Analytical Coverage: Detect 1,500+ glycerolipid species in a single run, enabling comprehensive lipidome profiling.
- High Quantitative Precision: Coefficient of variation (CV) ≤ 15% across technical replicates for reliable reproducibility.
- Ultra-Low Detection Limits: Limit of detection (LOD) down to 5 pg on column, suitable for trace-level lipid detection.
- Wide Dynamic Range: Accurate quantification across 10 pg – 10 μg for triacylglycerols and related species.
- Mass Accuracy You Can Trust: Orbitrap-based detection with < 2 ppm mass error, ensuring confident lipid identification.
Workflow for Glycerolipids Analysis Service
1. Sample Reception & Quality Check
Verify sample integrity and storage conditions. Assess matrix type (serum, tissue, plant extract, oils) and prepare for extraction.
2. Lipid Extraction & Cleanup
Employ modified Bligh & Dyer or MTBE-based extraction protocols. Remove interfering substances and ensure consistent lipid recovery.
3. Internal Standard Addition
Spike samples with isotope-labeled standards for each major glycerolipid class to ensure precise quantitation.
4. Chromatographic Separation (UHPLC)
Utilize Thermo Vanquish™ UHPLC with sub-2 μm C18 column for high-resolution separation of TAGs, DAGs, MAGs, and glycolipids. 18-minute gradient optimized for both neutral and polar lipids.
5. High-Resolution Mass Spectrometry
Perform analysis on Q Exactive™ HF-X Orbitrap or SCIEX TripleTOF® 6600+. Acquire full MS and MS/MS data for confident identification and quantification.
6. Data Processing & Annotation
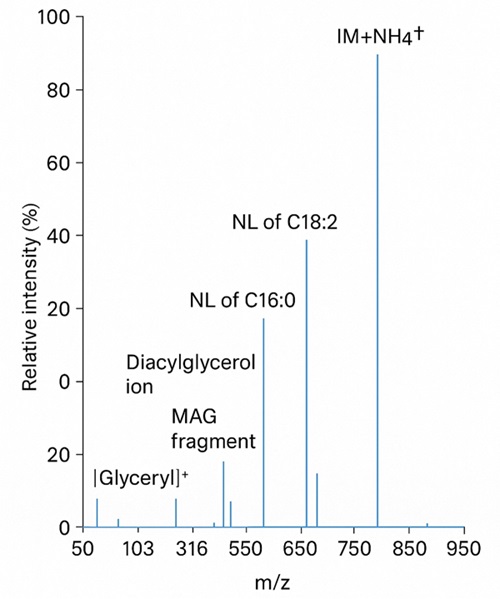
Process raw files with LipidSearch™, MS-DIAL, and in-house pipelines. Annotate species with accurate m/z, retention time, and MS/MS fragment data.
7. Statistical & Pathway Analysis
Provide differential analysis, heatmaps, PCA plots, and integration with KEGG/LipidMaps pathways.

Technology Platform for Glycerolipids Analysis Service
Chromatographic Separation (UHPLC)
- Instrument: Thermo Scientific™ Vanquish™ UHPLC
- Column: C18 reverse-phase, sub-2 μm particle size for sharp separation of TAG, DAG, MAG, and glycolipids
- Gradient: Optimized 18-min binary solvent system for both neutral and polar glycerolipids
- Flow Rate: 0.35 mL/min
- Carry-over: <0.05%, ensuring clean baselines for low-abundance species
 Vanquish™ UHPLC (Figure from Thermo Scientific)
Vanquish™ UHPLC (Figure from Thermo Scientific)
Mass Spectrometry Platforms
Orbitrap-Based High-Resolution Analysis
- Model: Thermo Scientific™ Q Exactive™ HF-X
- Ionization: Heated Electrospray Ionization (HESI) source, 2.8 kV
- Full MS: Resolution 240,000 at m/z 200; AGC target 3 × 10⁶; mass accuracy <2 ppm
- Data-Dependent MS/MS: Resolution 15,000; stepped collision energy for comprehensive fragmentation
- Polarity: Positive/negative switching for broad lipid class detection
 Q Exactive™ HF-X (Figure from Thermo Scientifi)
Q Exactive™ HF-X (Figure from Thermo Scientifi)
DIA Workflow for Comprehensive Coverage
- Model: SCIEX TripleTOF® 6500+
- Acquisition Mode: SWATH® DIA with 75 variable windows
- Cycle Time: 0.54 s for high-throughput screening
- Dynamic Range: 5 orders of magnitude for both abundant and trace lipids
 SCIEX Triple Quad™ 6500+ (Figure from Sciex)
SCIEX Triple Quad™ 6500+ (Figure from Sciex)
Sample Requirements for Glycerolipids Analysis Service
| Sample Type | Recommended Amount | Container | Storage Condition |
|---|---|---|---|
| Serum / Plasma | ≥ 200 µL | Cryovial (screw-cap) | -80 °C; avoid freeze–thaw |
| Tissue (animal / plant) | ≥ 50 mg (fresh or frozen) | Pre-labeled cryotube | Snap-frozen in liquid N₂; store at -80 °C |
| Cell Pellets | ≥ 1 × 10⁶ cells | Cryovial | Pellet washed with PBS; freeze immediately |
| Microbial Biomass | ≥ 50 mg (wet weight) | Sterile tube | Freeze-dried or snap-frozen |
| Plant Extract / Oils | ≥ 500 µL | Amber vial with Teflon cap | Protect from light; store at -20 °C or lower |
Notes:
- Avoid using EDTA or detergents as they can interfere with lipid extraction.
- Ensure samples are shipped on dry ice and remain frozen during transit.
- Contact us for specific guidelines on complex matrices or custom projects.
Demo Results
FAQ of Glycerolipids Analysis Service
Can you analyze both neutral glycerolipids and polar glycolipids in the same workflow?
Yes. Our LC–MS/MS platform resolves both neutral lipids (TAG, DAG, MAG) and polar glycolipids (MGDG, DGDG, SQDG) within one optimized chromatographic method, ensuring comprehensive coverage.
Do you provide absolute quantification, and how is it achieved?
We use isotope-labeled internal standards spanning major glycerolipid classes to provide absolute quantitation with calibration curves (R² ≥ 0.995), ensuring high accuracy.
How do you ensure structural confirmation for isomers and complex species?
Isomer discrimination is performed through advanced MS/MS fragmentation combined with retention-time prediction. For selected lipids, we apply diagnostic fragment ion analysis for precise identification.
What types of bioinformatics and statistical analyses are included?
Our deliverables feature lipid annotation, quantitative tables, lipid class summaries, PCA plots, volcano plots, heatmaps, and pathway interpretation integrated with KEGG and LipidMaps.
How do you maintain reproducibility across large sample sets?
We include QC samples in every batch, monitor retention time shifts, and apply stringent thresholds for mass accuracy (< 2 ppm) and CV (≤ 15%) to maintain consistency across large datasets.
Can you support high-throughput studies or large-scale lipid profiling projects?
Yes. Our platform is optimized for scalability with automated sample processing and DIA-based acquisition strategies (e.g., SWATH), enabling reliable data generation for large cohorts.
Is it possible to include rare or customized glycerolipid subclasses in the analysis?
Absolutely. We can build custom panels for rare lipid subclasses (e.g., betaine lipids, acylated galactolipids) and incorporate reference standards for accurate detection.
How do you manage matrix-specific challenges, such as plant glycolipids or oil-rich samples?
We adjust extraction protocols (e.g., MTBE, Bligh & Dyer variations) to ensure optimal recovery and apply matrix-specific cleanup strategies to minimize ion suppression.
Do you provide normalization strategies for comparative studies?
Yes. We offer multiple normalization options, including internal standard correction, total lipid signal normalization, and sample weight or protein content-based adjustment.
Can your data be integrated into multi-omics frameworks?
Yes. We provide standardized formats compatible with metabolomics and transcriptomics pipelines, enabling combined pathway modeling and network-level interpretation.
What measures do you take to control oxidation or degradation of sensitive glycerolipids?
We recommend cold-chain handling and add antioxidants when necessary. All extractions are performed under controlled conditions to prevent oxidative artifacts.
Learn about other Q&A about proteomics technology.
Publications
Here are some of the lipidomics-related papers published by our clients:

- White matter lipid alterations during aging in the rhesus monkey brain. 2024.
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024.
- Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. 2023.
- Lipid Membrane Engineering for Biotechnology (Doctoral dissertation, Aston University). 2023.
- Summative and ultimate analysis of live leaves from southern US forest plants for use in fire modeling. 2020.