Title: Profiling nucleotides in low numbers of mammalian cells by sheathless CE–MS in positive ion mode: Circumventing corona discharge
Journal: Electrophoresis
Published: 2020
Background
In metabolomics, the profiling of polar and charged metabolites, especially nucleotides, remains a challenge. These metabolites are crucial for cell signaling and metabolism, yet they tend to interact with stainless steel in LC–MS systems, which complicates analysis. An efficient microscale separation technique, such as capillary zone electrophoresis-mass spectrometry (CZE–MS), is required for nucleotide profiling in limited biological samples. CZE is well-suited for this task, offering high sensitivity with small sample volumes. Previous methods faced issues like electrolysis-induced corrosion and ionization suppression. The present study aims to overcome these challenges by using a sheathless porous tip interface and detecting nucleotides in positive ion mode, enabling the analysis of low numbers of mammalian cells and other metabolites.
Materials & Methods
Chemicals and Reagents
Various chemicals were used in the study, including ammonium acetate (99%), Dulbecco's PBS, DMEM/F-12, ammonium bicarbonate (99.5%), and ammonium carbonate, sourced from Sigma-Aldrich, Fluka, Scharlau, and Merck. Other chemicals like sodium hydroxide, hydrochloric acid, and ammonia were used to adjust pH, and reagents for mass spectrometry analysis included methanol, chloroform, and acetic acid. The water used was purified via a Milli-Q® Advantage A10 system. Fetal calf serum, penicillin-streptomycin, and centrifugal filters were also used for cell culture and sample processing.
Preparation of Solutions
Stock solutions of ammonium acetate, ammonium bicarbonate, and ammonium carbonate were prepared by dissolving them in water and adjusting the pH to specific values (8.5, 9.0, 9.7) using ammonia. Nucleotide stock solutions were prepared by dissolving compounds in a 95% water and 5% methanol mixture, and standards were mixed at a final concentration of 100 µM. Isotope-labeled nucleotides were used as internal standards for quantification, and aliquots were stored at −80°C.
Sample Preparation
To prepare the samples for analysis, chloroform saturated with water and methanol was used to extract nucleotides from biological samples. Calibration solutions were made by diluting stock nucleotide solutions. The internal standard solution was prepared with labeled nucleotides at 400 ng/mL concentration. The extraction involved a chloroform-methanol-water mixture followed by vortexing, centrifugation, and solvent evaporation using a SpeedVac concentrator. The final residues were reconstituted in a methanol/water mixture before MS analysis.
Cell Lysate Preparation
HepG2 cells were cultured and harvested, with a final cell density of 7.4 × 10⁶ cells/mL. Cells were cultured in Petri dishes and treated with ice-cold methanol/water solution to quench enzymatic activity. After centrifugation and filtration, the lysate was stored at −80°C. The lysates were then diluted to various cell densities (10,000 to 500 cells/50 µL) for further analysis.
CZE–MS Analysis
Capillary zone electrophoresis (CZE) was coupled with mass spectrometry (MS) for the analysis of nucleotides. A 30 µm i.d. fused-silica capillary was used for separation, with positive ionization mode in a Sciex TripleTOF 6600 MS system. The sheathless interface involved the use of a porous tip, and the sample was injected hydrodynamically. The MS parameters were optimized for ionization voltage, curtain gas pressure, and collision energy to ensure efficient nucleotide detection.
Determination of Analytical Performance
Calibration curves were generated by plotting the peak area ratios of nucleotides to internal standards against their concentrations. Limit of detection (LOD) was determined using a signal-to-noise ratio of 3, and accuracy was assessed by comparing calculated nucleotide concentrations with known values. The method's repeatability was evaluated by analyzing nucleotide samples from cell lysates prepared from different cell densities, ensuring consistency in results.
Results
Development of a Sheathless CZE–MS Method for Nucleotide Profiling
A highly sensitive sheathless capillary zone electrophoresis-mass spectrometry (CZE–MS) method was developed to profile nucleotides under high-pH separation conditions using ESI-TOF-MS in positive ion mode. This approach successfully minimized corona discharge and optimized conditions for nucleotide detection. Ammonium acetate at 16 mM and pH 9.7 was identified as the optimal background electrolyte (BGE), providing narrow peak shapes, high resolution, and acceptable CE currents. The use of methanol/water (1:1, v/v) for sample preparation enhanced signal intensities and signal-to-noise ratios.
Analytical Performance Evaluation
The method demonstrated excellent linearity (R² > 0.994), with limits of detection (LODs) ranging from 0.06 to 1.33 nM. Compared to existing methods such as ion-pair LC-MS and HILIC-MS, the developed sheathless CZE–MS exhibited significantly lower LODs, indicating superior sensitivity. Reproducibility was also high, with intraday and interday relative standard deviations (RSDs) for migration times and peak areas consistently below 15%. The method demonstrated good accuracy (90.7–102.4%) and negligible matrix effects, highlighting its robustness for nucleotide analysis in biological samples.
Profiling Nucleotides in Low Numbers of Mammalian Cells
The method was applied to HepG2 cell lysates, enabling nucleotide profiling from as few as 500 cells. Field-amplified sample stacking and transient-isotachophoresis (t-ITP) further improved sensitivity, achieving signal-to-noise ratio increases of up to 6.21-fold for AMP, ADP, and ATP. Good repeatability was observed for analyses using both 1000 and 2500 cells, with intra-dish CVs below 12%.
Broader Applicability
Beyond nucleotides, additional metabolites, including amino acids (e.g., proline, glutamic acid) and cofactors (e.g., NAD+, NADH, FAD), were detected, although further optimization is required to enhance sensitivity for certain compounds.
The results collectively demonstrate that the proposed sheathless CZE–MS method provides unparalleled sensitivity, precision, and reproducibility for profiling nucleotides and other metabolites in limited biological samples.
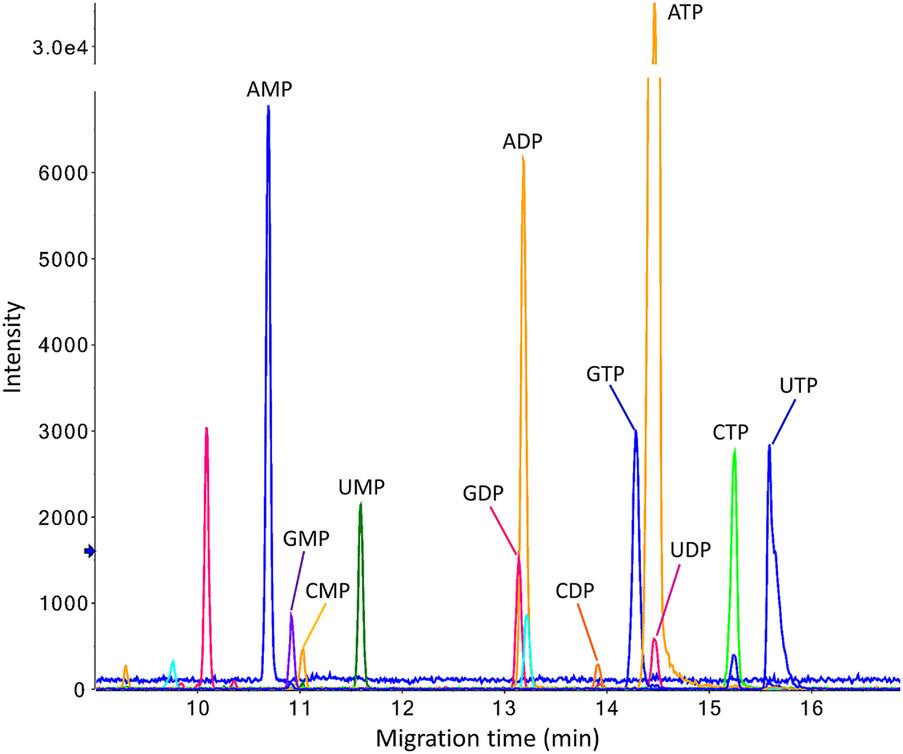
 Multiple extracted ion electropherograms obtained for the analysis of a standard nucleotide mixture (100 nM) by sheathless CE-MS in positive ion mode.
Multiple extracted ion electropherograms obtained for the analysis of a standard nucleotide mixture (100 nM) by sheathless CE-MS in positive ion mode.
 Reconstructed ion electropherograms obtained for the analysis of nucleotides in an extract from 10 000 HepG2 cells as starting material by sheathless CZE–MS in positive ion mode using a porous tip emitter.
Reconstructed ion electropherograms obtained for the analysis of nucleotides in an extract from 10 000 HepG2 cells as starting material by sheathless CZE–MS in positive ion mode using a porous tip emitter.
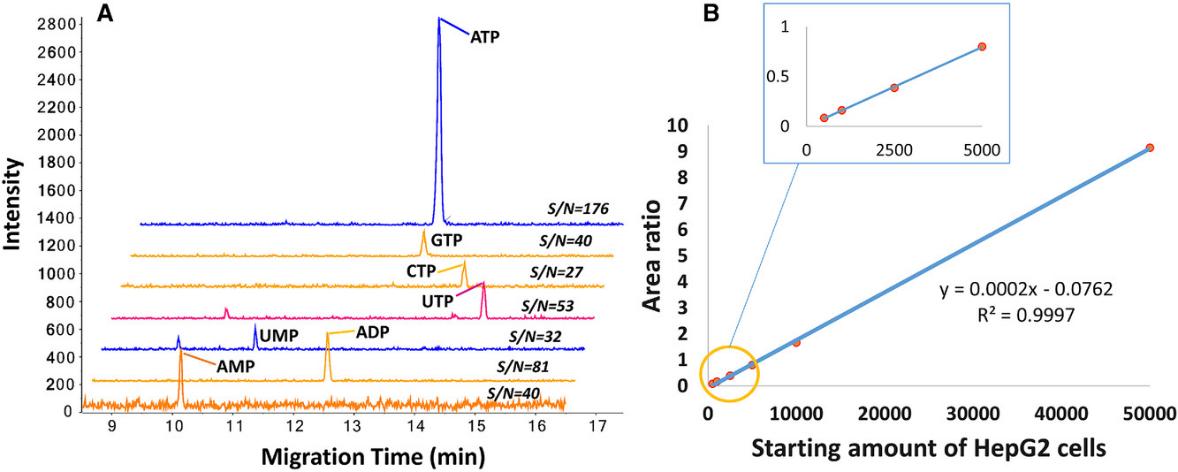 (A) Extracted ion electropherograms of nucleotides from 500 HepG2 cells analyzed by sheathless CZE–MS in positive ion mode. Conditions: 16 mM ammonium acetate (pH 9.7), 30 kV, 2.0 psi, 30 s. (B) Scatter plot of endogenous ATP/ISTD ratios vs. HepG2 cell numbers.
(A) Extracted ion electropherograms of nucleotides from 500 HepG2 cells analyzed by sheathless CZE–MS in positive ion mode. Conditions: 16 mM ammonium acetate (pH 9.7), 30 kV, 2.0 psi, 30 s. (B) Scatter plot of endogenous ATP/ISTD ratios vs. HepG2 cell numbers.
Reference
- Zhang, Wei, et al. "Profiling nucleotides in low numbers of mammalian cells by sheathless CE–MS in positive ion mode: circumventing corona discharge." Electrophoresis 41.5-6 (2020): 360-369. https://doi.org/10.1002/elps.201900417




