Redox Proteomics Service
Creative Proteomics provides professional Redox Proteomics research services, focusing on the analysis of the dynamic changes of proteins in the REDOX state and their biological functions. Our services cover key modifications such as thiol oxidation (e.g. S-nitrosylation, S-glutathione), disulfide bond formation, and protein carbonylation, combined with high-resolution LC-MS/MS and specific enrichment techniques for precise identification and quantitative analysis of modified proteins. In addition, we provide bioinformatic analyses that provide insight into the role of redox modifications in cell signaling, metabolic regulation, aging, and diseases such as cancer and neurodegenerative diseases. With advanced experimental platforms and customized research protocols, we enable researchers to fully explore the mechanisms of redox regulation and advance biomedical research.
Submit Your Request Now
×- What We Provide
- Technology Platforms
- Workflow
- Advantages
- Sample Requirements
- Applications
- Demo
- FAQ
- Publications
What Is Redox Proteomics?
Redox proteomics focuses on studying the modifications and dynamic changes of proteins during redox reactions[1]. Among the most common redox modifications are the oxidation of thiol groups (-SH) in cysteine residues to form disulfide bonds (-S-S-) or mixed disulfide adducts (-SSG), as well as the oxidation of other amino acid residues, such as tyrosine and methionine. According to the different redox reactions, common redox modifications include S-nitrosylation, S-sulfination, S-glutathionylation, S-thiothiolate, intermolecular and intramolecular disulfide bonds, S-acylation. By systematically analyzing the modifications at these redox-sensitive sites, one can not only uncover the potential impact of oxidative stress on protein function but also further explore the fine-tuned regulatory mechanisms of intracellular redox homeostasis, which has important clinical significance in neurodegenerative diseases such as cancer and Alzheimer's disease[2].
Redox Proteomics Service at Creative Proteomics
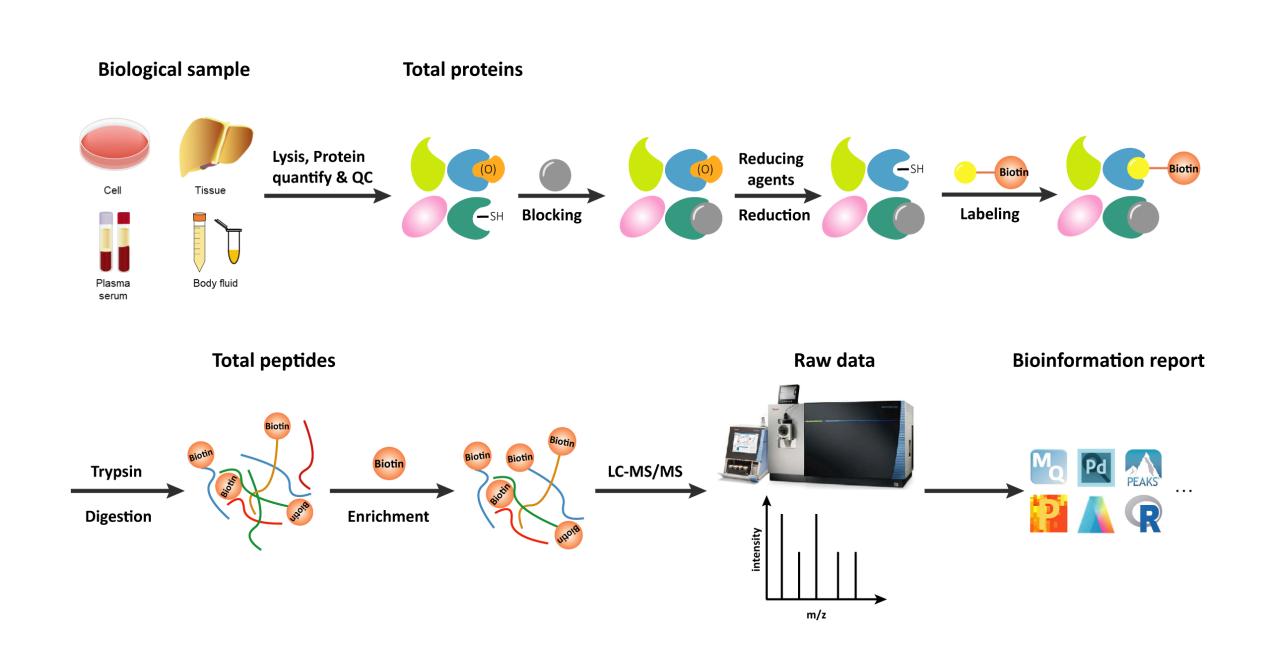
At Creative Proteomics, we have meticulously established and optimized our LC-MS/MS protocols to deliver a rapid, sensitive, and highly reliable site mapping service specifically designed for redox proteomics. Our advanced analytical platform integrates a high-performance C18 column with advanced instrumentation, including the Thermo Fisher Easy-nLC 1000 and the Thermo Fisher LTQ Orbitrap ETD. This cutting-edge setup allows us to achieve precise separation and high-resolution detection of redox modifications, enabling accurate identification and quantification of oxidative changes on proteins. Our comprehensive workflow covers every step—from robust sample preparation to detailed mass spectrometric analysis—ensuring that even subtle redox alterations are captured reliably.
Redox proteomics Research Platforms

Easy-nLC 1000 (Figure from Thermo Scientific)

Thermo Fisher LTQ Obitrap ETD (Figure from Thermo Scientific)
Workflow of Redox Proteomics Analysis
Advantages of Our Redox Proteomics Service
- Professional detection and analysis capability: Experienced research team, strict and matured techniques.
- Breadth: We could skillfully deal with protein samples derived from a wide range of sources, encompassing animal and plant tissues, bacteria, blood, membrane proteins, etc.
- High specificity and accuracy: Skillful quantification proteomics techniques, and PTMs enrichment methods.
- High stability and reproducible: Obtain consistent and reproducible results for data analysis.
- Advanced analytical platforms: Use of high-sensitivity mass spectrometry and advanced tools, ensuring precise and repeatable results.
- End-to-End Support: Offer full-spectrum services from design to data interpretation, with continuous technical support throughout the research process.
Redox Service Sample Requirements
| Sample Type | Sample Quantity | |
|---|---|---|
| Tissue | animal tissue | 200-500mg |
| fresh plant | 200-500mg | |
| Cell | suspension cell | > 3x107 |
| adherent cell | > 3x107 | |
| microorganism | > 50 mg or 3 × 107 cells | |
| Body fluid | Serum/plasma | 1mL |
| Protein | Total protein > 3mg and concentration >1 μg/μL | |
Applications of Redox Proteomics Service




Biomarker discovery
Specific oxidation-modified proteins can be used as potential biomarkers for disease diagnosis and prognosis.
Drug action mechanism and target discovery
Evaluate how drugs affect cellular oxidative modification networks, reveal their mechanisms of action, and discover new therapeutic targets to aid drug development.
Mechanism of oxidative stress and disease[2]
Systematically analyze how oxidative modification of proteins affects protein function, thereby revealing molecular mechanisms such as cancer, neurodegenerative diseases, and cardiovascular diseases.
Plant and agricultural research[1]
Analysis of oxidative modification patterns of plants can optimize the study of crop stress resistance.
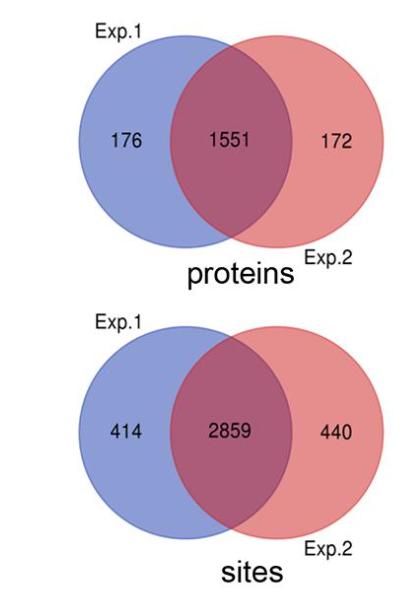
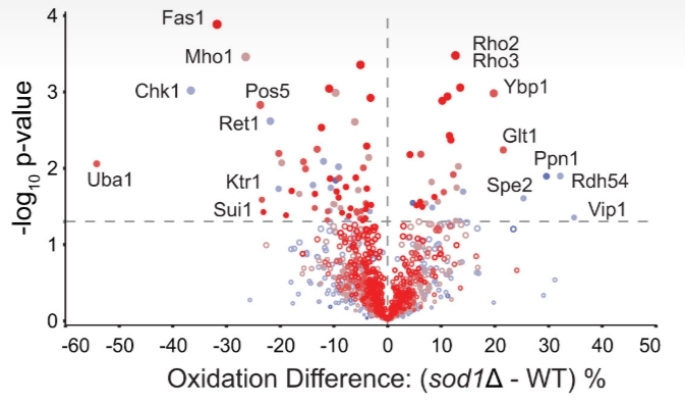
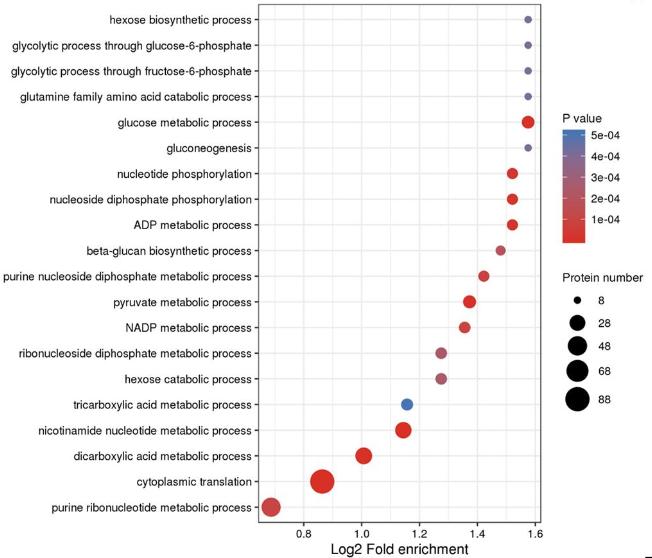
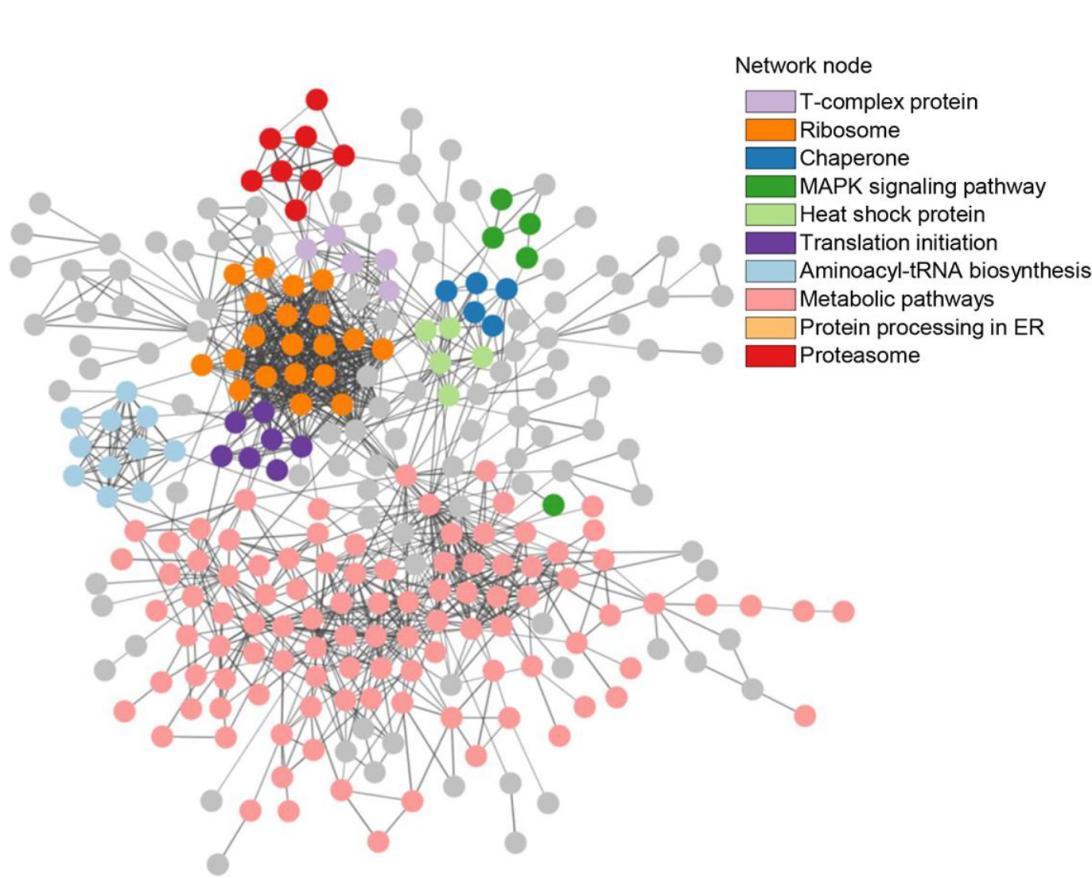
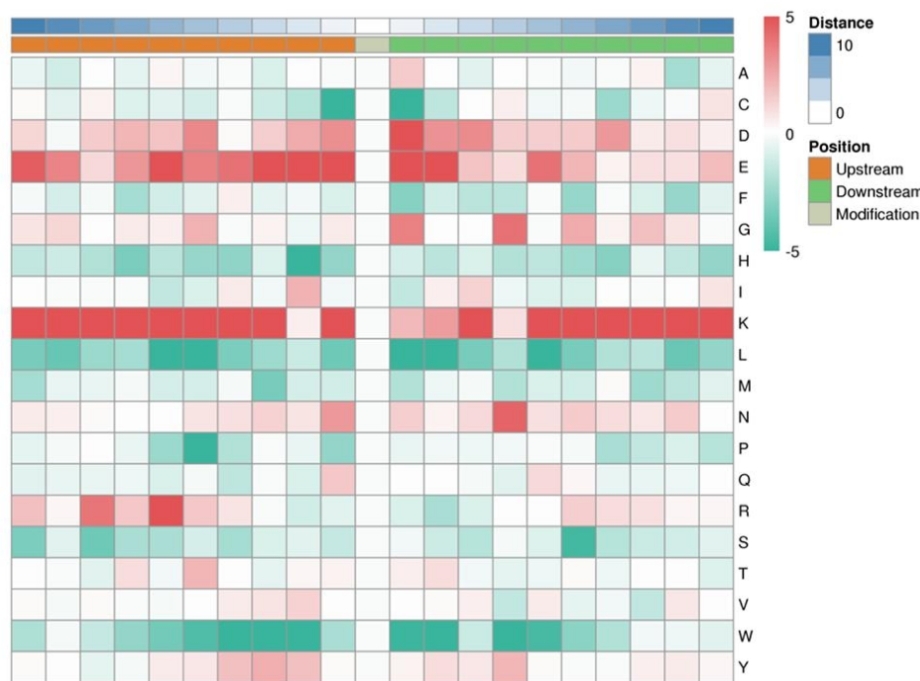
Demo Result of Redox Proteomics Service
FAQs of Redox Proteomics Service
How to ensure the stability of the REDOX state?
Reducing agents (such as DTT, TCEP) and alkylating agents (such as iodoacetamide) are used during sample processing to stabilize the REDOX state of cysteine, and experimental conditions are strictly controlled to avoid anthropogenic oxidation.
Can REDOX modifications be quantitatively analyzed?
Yes, mass spectrometry-based Label-free can be employed to accurately measure changes in REDOX modifications and provide quantitative results.
Which sample types are accepted by the service?
We accept a variety of sample types including cells, tissues, blood, serum, plasma, plant tissue and more. If you have other special sample types, please consult in advance.
Whether the sample needs pretreatment?
We recommend that customers provide fresh or frozen samples, which should avoid repeated freezing and thawing, and we will be responsible for subsequent protein extraction and pretreatment. If you have already extracted the protein, please communicate with us in advance about the sample handling method.
Learn about other Q&A.
Redox Proteomic Case Study
Publications
Here are some publications in Proteomics research from our clients:

- APOB100 transgenic mice exemplify how the systemic circulation content may affect the retina without altering retinal cholesterol input. 2024. https://doi.org/10.1167/iovs.61.13.19
- Hyperfunction of post-synaptic density protein 95 promotes seizure response in early-stage aβ pathology. 2024. https://doi.org/10.1038/s44319-024-00090-0
- Confirmation of translatability and functionality certifies the dual endothelin1/VEGFsp receptor (DEspR) protein. 2016. https://doi.org/10.1186/s12867-016-0066-8
- Molecular signature of the ontogenic development of the prawn Macrobrachium tenellum. 2023. https://doi.org/10.7717/peerj.16344
- In vitro assays reveal inherently insecticide-tolerant termite symbionts. 2023. https://doi.org/10.3389/fphys.2023.1134936
References
- Ramakrishnan, M., et al. "Redox status of the plant cell determines epigenetic modifications under abiotic stress conditions and during developmental processes." Journal of advanced research, (2022): 42, 99–116. https://doi.org/10.1016/j.jare.2022.04.007
- Cadenas-Garrido P., et al. "Using Redox Proteomics to Gain New Insights into Neurodegenerative Disease and Protein Modification." Antioxidants (Basel, Switzerland), (2024): 13(1), 127. https://doi.org/10.3390/antiox13010127
- Zhang, X., et al. "The Redox Proteome of Thiol Proteins in the Rice Blast Fungus Magnaporthe oryzae." Frontiers in microbiology, (2021): 12, 648894. https://doi.org/10.3389/fmicb.2021.648894
- Montllor-Albalate, C., et al. Sod1 integrates oxygen availability to redox regulate NADPH production and the thiol redoxome. Proceedings of the National Academy of Sciences of the United States of America, (2022): 119(1), e2023328119. https://doi.org/10.1073/pnas.2023328119