Introduction to Protein Identification and Tandem Mass Spectrometry
In modern proteomics, protein identification is fundamental for advancing our understanding of biological systems, disease mechanisms, and therapeutic targets. Tandem Mass Spectrometry (MS/MS) has revolutionized the process of protein identification and analysis by offering precise and high-throughput capabilities to analyze complex protein mixtures with remarkable sensitivity. The purpose of this article is to explore how tandem mass spectrometry is applied in protein identification, specifically discussing LC-MS/MS methodologies, workflows, challenges, and applications across various biological and clinical fields.
The identification and characterization of proteins are central to life sciences, enabling us to unravel the molecular mechanisms of cellular functions, disease states, and responses to drugs. Mass spectrometry (MS), and more specifically tandem mass spectrometry (MS/MS), has emerged as the premier technique for protein identification due to its accuracy and ability to analyze large datasets rapidly. By coupling liquid chromatography (LC) with tandem MS, researchers are able to achieve even greater separation and analysis of proteins in complex biological samples—a process referred to as LC-MS/MS protein identification.
Why Mass Spectrometry for Protein Identification?
Traditional methods of protein identification often fall short when handling complex mixtures or low-abundance proteins. Protein mass spectrometry has enabled researchers to bypass these limitations by identifying and characterizing proteins based on their mass-to-charge (m/z) ratio. Protein mass spec can rapidly analyze mixtures, making it possible to conduct protein identification and protein analysis on a large scale.
By combining liquid chromatography with tandem mass spectrometry, LC-MS/MS enables efficient separation of peptides before they enter the mass spectrometer. This approach, known as LC-MS/MS tandem mass spectrometry, is particularly valuable in protein analysis by mass spectrometry, as it ensures that each peptide is analyzed with minimal interference from other peptides in the mixture.
Services you may be interested in:
Principles of Tandem Mass Spectrometry for Protein Identification
Tandem mass spectrometry (MS/MS), also known as tandem MS or MS-MS, operates through a two-stage process designed to precisely analyze complex proteins or peptides by fragmenting and sequencing their components. This technique is especially advantageous for protein identification because it enables the detailed breakdown of proteins into smaller, identifiable peptides, which can then be matched to known sequences in protein databases.
Ionization: Converting Proteins to Charged Particles
The first essential step in tandem mass spectrometry is ionization, a process that converts proteins or peptides into gas-phase ions so they can be manipulated and measured by the mass spectrometer. Two of the most common ionization methods used in protein mass spectrometry are:
- Electrospray Ionization (ESI): In ESI, the protein sample is introduced in a liquid form and passes through a fine needle under high voltage. This process produces a fine mist of charged droplets, from which solvent evaporates, leaving behind charged protein or peptide ions.
- Matrix-Assisted Laser Desorption/Ionization (MALDI): Here, the protein sample is embedded in a crystalline matrix and subjected to a laser, which vaporizes the matrix and ionizes the protein. MALDI is particularly suited for high-mass proteins and is often used in protein analysis applications requiring broad mass ranges.
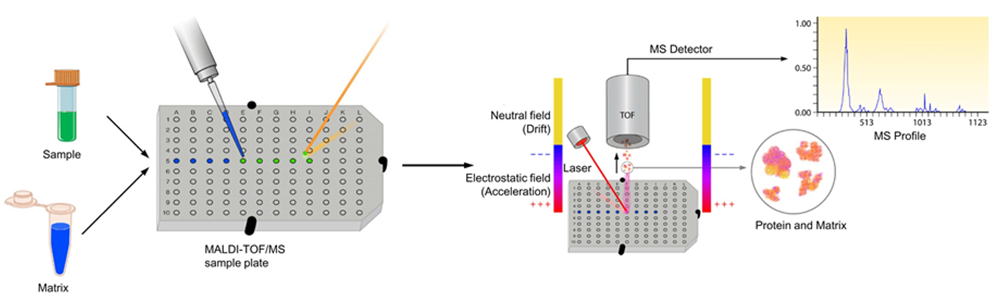 Figure 1. The process of MALDI-TOF mass spectrometry
Figure 1. The process of MALDI-TOF mass spectrometry
These ionization techniques enable mass spectrometry for protein analysis by creating ions that can be measured based on their mass-to-charge (m/z) ratios.
Mass Selection: Isolating Specific Ions
After ionization, the resulting ions are directed into the first mass analyzer. In tandem MS, this analyzer functions as a filter, allowing only ions of a specific m/z ratio to pass through to the next stage. This selection step is essential for protein identification by mass spectrometry as it isolates a particular ion for detailed analysis without interference from other ions present in the sample.
There are several types of mass analyzers used in tandem mass spectrometry, each with unique characteristics:
- Quadrupole Mass Analyzers: Use electric fields to filter ions by their m/z ratio. Commonly employed in tandem MS workflows, quadrupoles are highly effective for isolating specific ions and are often used in LC-MS/MS tandem mass spectrometry.
- Time-of-Flight (TOF) Analyzers: Measure the time it takes for ions to reach the detector, with lighter ions arriving faster than heavier ones. TOF analyzers provide high resolution, making them suitable for complex protein mixtures.
- Orbitrap and Fourier Transform Ion Cyclotron Resonance (FTICR): These high-resolution analyzers are ideal for precision protein analysis by mass spectrometry and can detect very small differences in mass, which is critical in identifying and distinguishing proteins.
Fragmentation: Breaking Down Peptides for Sequencing
Once isolated, the selected ions undergo fragmentation in a collision cell, where they are bombarded with inert gas molecules (usually nitrogen or helium). This step, known as collision-induced dissociation (CID) or higher-energy collisional dissociation (HCD), breaks the peptide ions into smaller fragments, typically at peptide bonds. Fragmentation generates a series of ions that reflect the structure and sequence of the original peptide, a feature critical for peptide identification by mass spectrometry.
Fragment ions are typically categorized as b-ions and y-ions, representing fragments where the charge remains on the N-terminus or C-terminus of the peptide, respectively. The pattern of these ions across the MS/MS spectrum forms a unique fingerprint that can be mapped to peptide sequences within a database, a core step in mass spectrometry protein identification.
Second Mass Analysis and Detection
The fragmented ions produced in the collision cell then enter the second mass analyzer, where their m/z ratios are measured and recorded, generating the final MS/MS spectrum. This MS/MS spectrum provides a comprehensive view of the peptide's fragmentation pattern, which is essential for determining the peptide's amino acid sequence. The level of detail in this spectral data is crucial for identification of proteins by mass spectrometry and supports accurate protein characterization, including the identification of post-translational modifications (PTMs).
Data Interpretation and Matching with Protein Databases
The MS/MS spectrum is subsequently analyzed using bioinformatics tools, which compare the fragment patterns to a protein database. Algorithms such as Mascot, SEQUEST, and MaxQuant facilitate protein identification by scoring matches between the observed spectra and theoretical peptide spectra generated from known protein sequences. This matching process allows researchers to pinpoint the protein's identity with high confidence and, in many cases, to characterize specific modifications or structural features.
How to Identify Proteins by LC-MS/MS?
1) Protein Sample Preparation
Sample preparation begins with protein extraction and purification from the biological matrix. Samples are often enzymatically digested (commonly with trypsin) into smaller peptides to improve detection and identification efficiency.
2) Liquid Chromatography (LC) Separation
In LC-MS/MS for protein identification, peptides are first separated by liquid chromatography. The LC stage reduces sample complexity, allowing each peptide to enter the tandem MS system with minimal overlap, which enhances the reliability of protein identification using mass spectrometry.
3) Tandem Mass Spectrometry Analysis
During the MS stage, peptides are ionized and filtered through the first mass analyzer, which selects ions for fragmentation in the collision cell. The resultant fragment ions are detected in the second mass analyzer, generating spectra that reveal the peptide's sequence. This sequence data is critical in identification of proteins by mass spectrometry as it enables researchers to reconstruct the primary structure of the peptides.
4) LC-MS/MS Data Acquisition
In the data acquisition phase, the tandem mass spectrometer collects raw spectral data, including mass-to-charge (m/z) ratios of peptide ions and their fragment ions, along with their intensity values. This data acquisition process is time-synchronized with the LC separation, so the system can correlate specific peptides with retention times, adding another layer of specificity to protein mass spectrometry. The acquired MS/MS data is stored in a raw format for further computational analysis, where the spectra will be matched against protein databases to facilitate protein identification.
5) Data Analysis for Protein Identification
The identification process culminates in bioinformatics. Specialized software, such as Mascot, SEQUEST, and MaxQuant, compares the MS/MS spectra against known protein databases, enabling identification of proteins with a high degree of accuracy. Peptide identification by mass spectrometry is pivotal here, as it allows matching of individual peptides to the target protein.
 Figure 2. Schema of a tandem mass spectrometry associated with liquid chromatography (LC-MS/MS)
Figure 2. Schema of a tandem mass spectrometry associated with liquid chromatography (LC-MS/MS)
Advantages of Tandem MS in Protein Identification
High Specificity through Fragmentation Analysis
One of the most valuable features of tandem MS is its ability to conduct fragmentation analysis. By breaking down peptides into smaller fragments, tandem MS produces a unique spectrum that reflects the sequence of amino acids within a peptide. This fragmentation yields b-ions and y-ions, which represent N-terminal and C-terminal fragments, respectively, and together provide a fingerprint specific to each peptide.
Fragmentation patterns allow tandem MS to distinguish between proteins with high precision, even among closely related variants or homologous sequences. This level of specificity is crucial when analyzing complex samples that contain thousands of proteins, as tandem MS can accurately identify peptides even in the presence of isobaric or similar peptides.
High Sensitivity for Low-Abundance Proteins
Protein mass spectrometry via tandem MS is highly sensitive, allowing for the detection of proteins present in very low concentrations. This sensitivity is particularly advantageous in proteomics, where many biologically significant proteins (e.g., signaling molecules, transcription factors, or biomarkers) may exist in low abundance and would be difficult to detect with less sensitive methods.
Using advanced ionization techniques such as electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI), tandem MS can identify and quantify proteins with minimal sample loss. This sensitivity enables researchers to achieve robust protein identification even in samples with challenging concentration ranges, such as blood plasma or cellular lysates.
High Throughput for Large-Scale Proteomics
In LC-MS/MS proteomics, the tandem MS setup coupled with liquid chromatography allows for high-throughput analysis of complex protein mixtures. The LC component enables separation of peptides before they reach the mass spectrometer, ensuring each peptide is independently analyzed with reduced signal interference from overlapping ions. This configuration accelerates the identification process, making tandem MS ideal for large-scale proteomic studies where hundreds or thousands of proteins may need to be identified in a single experiment.
High throughput is a significant advantage in research areas that require rapid data acquisition, such as biomarker discovery, drug development, and clinical diagnostics. With tandem MS, researchers can analyze multiple samples and conditions in a shorter time, obtaining a more comprehensive view of the proteome and generating data critical for biological insights.
Ability to Detect and Characterize Post-Translational Modifications (PTMs)
Post-translational modifications (PTMs) like phosphorylation, glycosylation, and ubiquitination are critical for protein function and regulation, yet challenging to identify due to their structural diversity and low abundance. Tandem MS excels in characterizing PTMs by capturing the precise fragmentation pattern of modified peptides, which is essential for identifying both the modification type and location on the protein.
Through targeted fragmentation, tandem MS can distinguish PTMs from unmodified peptide forms, generating unique ion patterns that signal the presence and specific placement of modifications. This capability is invaluable for studies that require detailed protein characterization and is widely used in mass spectrometry for protein analysis in functional proteomics.
Data Compatibility with Advanced Bioinformatics Tools
Another advantage of tandem MS is the compatibility of its data outputs with various bioinformatics platforms, which greatly enhances the ability to process and interpret complex MS/MS spectra. Protein identification by mass spectrometry relies on matching the acquired spectra against known protein databases, a task managed by software such as Mascot, SEQUEST, and MaxQuant.
These tools use algorithms to compare fragmentation patterns from tandem MS to theoretical spectra, facilitating efficient and accurate protein identification across a wide range of species and sample types. By integrating with such software, tandem MS enables researchers to leverage powerful database matching capabilities, which is crucial for identifying novel or unexpected proteins in large datasets.
Versatility across Different Biological Samples and Experimental Designs
Tandem MS is highly adaptable to a variety of sample types and experimental setups. Whether analyzing simple mixtures, complex biological fluids, or tissue samples, tandem MS can be optimized for specific protein analysis by mass spectrometry needs. The versatility of tandem MS also extends to different modes of quantification, including both label-free and labeled quantitation (e.g., SILAC, TMT), making it suitable for comparative proteomics studies that require precise measurement of protein abundance across multiple conditions.
This flexibility allows tandem MS to be applied broadly, from basic biological research to clinical and pharmacological applications, where it provides essential insights into protein structure, function, and interaction networks.
High Precision and Reproducibility in Protein Identification
The reproducibility of tandem MS is another key benefit. By using controlled ionization and fragmentation processes, tandem MS produces consistent, high-quality spectra that can be repeatedly analyzed to confirm protein identification. This precision is particularly valuable for studies requiring high-confidence data, such as those in regulated fields like clinical proteomics or therapeutic protein analysis.
References
- Raquel Pérez-Míguez, María Luisa Marina, María Castro-Puyana. High resolution liquid chromatography tandem mass spectrometry for the separation and identification of peptides in coffee silverskin protein hydrolysates. Microchemical Journal, 2019,149.
- Swearingen Kristian E, Eng Jimmy K, Shteynberg David, et al. A Tandem Mass Spectrometry Sequence Database Search Method for Identification of O-Fucosylated Proteins by Mass Spectrometry. Journal of proteome research, 2019,18(2).