What is Peptide Mass Fingerprinting (PMF)?
Peptide mass fingerprinting (PMF), also known as protein fingerprinting, is a high-throughput analytical method that developed in 1933 to identify proteins. Endoproteases first cleaves the unknown target protein into smaller peptides. The absolute mass of these peptides can be accurately measured using a mass spectrometer (such as MALDI-TOF), and a list of peptide peaks of the unknown protein is also obtained. This list of peaks is compared to a list of theoretical peptide peaks from a protein database using a computer program. This program translates all organism's known genomes into proteins, theoretically digests the proteins into small peptides, and calculates the absolute mass of the peptide from each protein. At the same time, the mass of the peptide from the lysed unknown protein is also compared with the theoretical peptide mass of each protein encoded in the genome. The results are statistically analyzed to find the best match.
The Process of Peptide Mass Fingerprinting
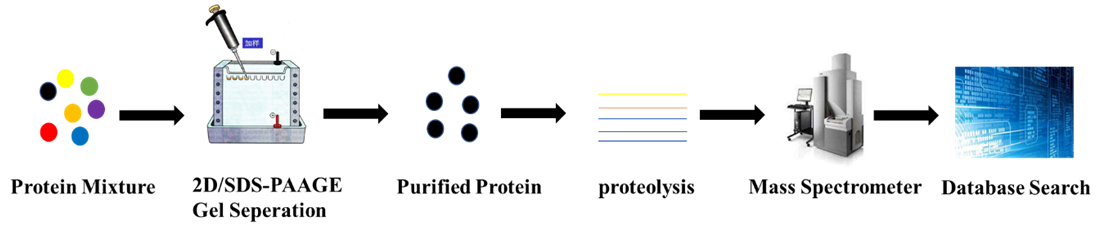
After the proteins in biological tissues or cells are purified, they are separated by 2D or SDS-PAGE gel and compared using imaging techniques. Proteins with significant differences are selected for further analysis. A series of peptide compounds that reflect the characteristics of the protein are obtained by degumming and enzymatic hydrolysis of the selected protein. The fingerprint of the peptide mixture is accurately measured by a mass spectrometer to obtain the molecular weight of the protein. Searching through a known database provides quasi-determinism in the selected protein.
The Applications of Peptide Mass Fingerprinting
With the development of analytical chemistry technology, PMF has been widely used in various fields such as drug authenticity identification, food quality control and human disease diagnosis due to its high sensitivity, high stability and low sample loss. The World Health Organization and the State Food and Drug Administration of China use PMF as a material identification and process control technology to provide objective indicators for quality evaluation.
Advantages and Disadvantages of Peptide Mass Fingerprinting
➢ Advantages
- Does not rely on protein sequencing for protein identification, but the peptide quality.
- Simple and fast, a classic method for the identification of proteins by first-order mass spectrometry
➢ Disadvantages
- The protein sequence of interest must exist in the target database.
- Good for single protein analysis. Presence of mixed proteins greatly increases the complexity of the analysis.
- Peptides of similar quality will greatly increase the difficulty of matching.
MALDI-TOF Peptide Mass Fingerprinting
Rapid and accurate protein identification is essential in proteomics research. PMF can be measured by matrix-assisted laser desorption ionization ion source (MALDI) and time-of-flight (TOF) mass analyzer. It is the most commonly used method in proteomics research.
MALDI uses a laser to irradiate a co-crystal thin film formed by a sample and a substrate. The substrate absorbs energy from the laser and transfers it to biomolecules. In the ionization process, the protons are transferred to or obtained from the biomolecules, and the biomolecules are ionized. Therefore, it is a soft ionization technique suitable for the determination of mixtures and biological macromolecules. The principle of TOF is that ions are accelerated to fly through the flight duct under the action of an electric field, and the ions are detected based on the time of flight to the detector. MALDI-TOF-MS has the characteristics of high sensitivity, high accuracy, and high resolution. It provides an increasingly important role for sample analysis in life sciences and many other fields.
References
- Cottrell J S. Protein identification by peptide mass fingerprinting. Peptide research, 1994, 7(3): 115-124.
- Hjernø K. Protein identification by peptide mass fingerprinting//Mass Spectrometry Data Analysis in Proteomics. Humana Press, 2007: 61-75.
- Suckau D, Resemann A, Schuerenberg M, et al. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Analytical and bioanalytical chemistry, 2003, 376(7): 952-965.