Co-immunoprecipitation (Co-IP) Analysis Service
Creative Proteomics offers comprehensive and fully customizable Co-Immunoprecipitation (Co-IP) services, including experimental design, antibody selection, buffer optimization, bead selection, sample preparation, protein capture and precipitation, washing, and elution. We employ advanced analysis techniques such as SDS-PAGE, Western Blot, and high-resolution mass spectrometry (Co-IP/MS) for in-depth protein interaction studies. Additionally, we provide detailed statistical and graphical analysis reports, supporting research on protein-protein interactions, signaling pathways, and disease mechanisms.
Submit Your Request Now
×- Define
- What We Provide
- How to Conduct
- Advantages
- IP vs. CoIP
- Sample Requirements
- Case
- FAQs
- Publications
What is Co-immunoprecipitation (Co-IP)?
The Co-IP technique is widely recognized as one of the most robust methods employed for investigating protein-protein interactions in vivo under non-denaturing conditions. These interacting proteins may be complex partners, structural proteins, co-factors, and signaling molecules. Co-IP as an enrichment method is also essential to identify low abundance target protein in a complex biological milieu. Following Co-IP, these protein complexes can be identified in various methods, such as Western Blot (WB) and LC-MS/MS, to identify partner proteins' ID, as well as exploration of undiscovered functions associated with bait protein. Additionally, quantitative proteomics such as SILAC, TMT/iTRAQ technologies can be equally applied in conjunction with Co-IP for precise quantification.
However, there poses a challenge due to the presence of highly abundant proteins and the vast array of proteins in cell lysates. Especially, this challenge arises from the inherent instability bindings, non-specific bindings, and potential antibodies contamination, which can potentially hinder the accurate detection of protein-protein interaction. Therefore, it is imperative to establish an appropriate negative control group in Co-IP experiments to minimize the risk of obtaining false positive results and ensuring the reliability of the obtained information on interacting protein.
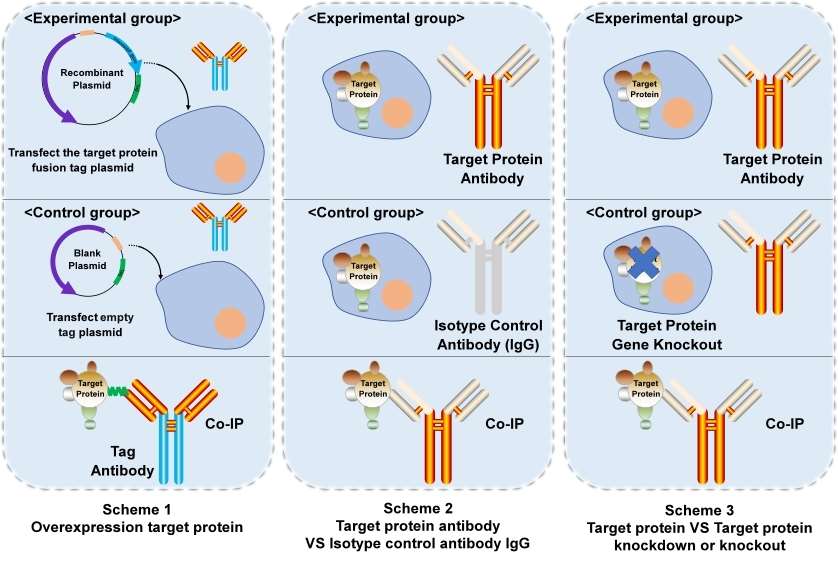 Figure 1. Three common schemes of Co-IP.
Figure 1. Three common schemes of Co-IP.
There are primarily three approaches for establishing negative control group: 1) Overexpression of the target protein using recombinant plasmid versus empty plasmid; 2) Utilization of target protein antibody versus Isotype nonfunctional control antibody; 3) Comparison between normal expression of the target protein and knockout or knockdown models.
Table 1. The advantages and disadvantages of each scheme.
| Scheme | Advantages | Disadvantages |
|---|---|---|
| 1 | 1) High protein expression; 2) Cost-effectiveness; 3) Well-established tag antibody system. | 1) Recombinant plasmids construction; 2) Target protein higher levels than physiological states. |
| 2 | 1) High confidence; 2) Proteins remain in their natural states. | 1) higher specific target protein antibody; 2) Low expression of target protein that may cannot be detected. |
| 3 | 1) High confidence; 2) Under natural condition. | 1) Time consuming; 2) Higher specific target protein antibody; |
Co-Immunoprecipitation Service in Creative Proteomics
Creative Proteomics provides a fully customizable Co-IP service that accommodates the unique requirements of each research project. With our in-depth expertise and advanced technology, we deliver comprehensive support through each phase of the Co-IP experiment, ensuring precise and reproducible results. Our Co-IP service includes:
Tailored Experimental Design
Each Co-IP experiment is adapted to fit the specific goals and conditions of your study:
- Antibody Selection: We provide an array of high-quality antibodies to target a wide range of proteins, or we can use tagged antibodies (e.g., Myc, HA, Flag) if the bait protein lacks a suitable specific antibody.
- Buffer Optimization: We customize lysis and wash buffers to suit your protein of interest, maximizing both yield and purity.
- Bead Selection: Choose between agarose or magnetic beads depending on the requirements of your protein interaction study.
Parameter Optimization for Reliable Results
Our team of experts optimizes critical parameters to enhance the specificity and efficiency of protein capture:
- Incubation Time: Adjusted to achieve optimal binding between the bait protein and its interaction partners.
- Elution Conditions: Calibrated to maximize recovery of protein complexes while minimizing non-specific binding and degradation.
Streamlined Experimental Workflow
Our Co-IP process follows a structured workflow, executed by experienced specialists to ensure accuracy at each step:
- Sample Collection and Preparation: We provide protocols for both endogenous and non-endogenous interaction studies. For endogenous interactions, protein samples are obtained by lysing co-transfected cells, while non-endogenous interactions are set up by combining purified bait proteins with cell or tissue lysates.
- Antibody Capture and Precipitation: We use antibody-conjugated beads to isolate the bait protein and its binding partners. If necessary, tagged antibodies can be employed to capture bait proteins when specific antibodies are unavailable.
- Washing and Elution: Thorough washing to reduce background noise, followed by careful elution to collect the protein complexes.
Advanced SDS-PAGE, Western Blot Analysis, and Mass Spectrometry (Co-IP/MS)
To confirm the presence of protein complexes:
- SDS-PAGE Separation: Protein complexes are separated to assess the molecular weights and identities of each component.
- Western Blot (WB) Verification: WB detection is used to verify the presence of specific target proteins in the precipitated complex, particularly when molecular weights are known.
- Co-IP/MS Analysis: For in-depth characterization, we utilize Co-IP/MS, combining co-immunoprecipitation with mass spectrometry to identify and quantify protein interaction partners at high resolution. This provides insights into complex protein networks and enables precise quantification of interacting proteins.
Comprehensive Data Analysis and Reporting
Following Co-IP, we conduct rigorous data analysis and provide a detailed report including:
- Thorough Mass Spectrometry Analysis: Using mass spectrometry (Co-IP/MS), our bioinformatics team performs statistical and graphical analysis of identified proteins, yielding high-resolution interaction data.
- Customizable Report Format: We provide data in formats tailored to meet your publication and presentation needs, allowing seamless integration of results.
How to Conduct Co-Immunoprecipitation at Creative Proteomics
1) Initial Consultation: Reach out to our team to discuss your specific research goals and requirements. We will help you define the scope of the Co-IP project.
2) Design Agreement: We will draft a detailed experimental design based on your feedback, outlining the selected antibodies, buffers, and other parameters.
3) Sample Preparation: You will provide cell lysates or we can assist in the preparation process, ensuring optimal conditions for Co-IP.
4) Execution of Co-IP: Our experienced technicians will carry out the Co-IP, adhering to strict protocols and quality control measures.
5) Analysis and Reporting: Once the Co-IP is complete, we will analyze the eluted proteins and deliver a comprehensive report detailing our findings.
Advantages of Co-Immunoprecipitation Assay
- Enhanced Sensitivity with Silver Staining: Our use of silver staining technology offers sensitivity up to 100 times greater than Coomassie Brilliant Blue staining, resulting in exceptionally clear SDS-PAGE electrophoresis results.
- Comprehensive Detection Equipment: Equipped with advanced liquid chromatography-mass spectrometry (LC-MS) and Fortebio systems, we provide a complete suite of analytical tools for further in-depth analysis of detection results.
- Increased Reliability with Repeated Experiments: Each experiment is conducted in duplicate to mitigate the impact of random variables, ensuring reliable and reproducible results.
- Integrated Protein and Antibody Preparation Platform: Our facilities include complete protein and antibody preparation services. Simply provide us with your sequence, and we will handle the entire experiment for you.
Choosing Between Immunoprecipitation (IP) and Co-Immunoprecipitation (Co-IP)
If You Need to Study a Single Protein:
Recommended Service: Immunoprecipitation (IP)
Why: IP is ideal if your objective is to isolate and analyze a specific protein, enabling you to focus on its structure or function without interference from other proteins.
Suggested Applications: Structural analysis, functional assays, antibody validation, or single-protein quantification.
If You're Investigating Protein-Protein Interactions or Complexes:
Recommended Service: Co-Immunoprecipitation (Co-IP)
Why: Co-IP is specifically designed to capture and identify proteins that interact with your protein of interest, helping you understand complex cellular mechanisms and signaling pathways.
Suggested Applications: Studies on cellular signaling, protein interaction networks, pathway discovery, or disease mechanism research.
For In-Depth Quantitative and Qualitative Analysis of Protein Complexes:
Recommended Service: Co-Immunoprecipitation with Mass Spectrometry (Co-IP/MS)
Why: Adding MS to Co-IP allows for high-resolution identification and quantification of each component within a protein complex, giving detailed insights into protein interaction partners and interaction strengths.
Suggested Applications: Proteomics studies, discovery of novel protein interactions, and quantitative analyses of complex cellular pathways.
Unsure Which Option is Best?
Contact Our Team for a Consultation: If you're uncertain which service aligns with your research goals, our experts are available to discuss your project requirements and recommend the best approach based on your objectives and expected outcomes.
Sample Requirements of Co Immunoprecipitation Assay
| Sample Type | Description | Required Volume |
|---|---|---|
| Cell Lysates | Lysates from cultured cells expressing the bait protein or co-transfected cells. | 200 µL - 1 mL |
| Tissue Homogenates | Homogenized tissues containing the protein of interest; should be freshly prepared. | 50 mg of tissue in 500 µL buffer (equivalent to 200 µL of lysate) |
| Purified Proteins | Recombinant proteins purified from expression systems; ideally tagged for easier isolation. | 50 - 200 µg |
| Serum or Plasma Samples | Human or animal serum/plasma; may contain endogenous proteins of interest. | 100 µL - 1 mL |
| Transfected Cell Lines | Cells transfected with plasmids expressing bait proteins. | 1 x 10^6 - 5 x 10^6 cells (equivalent to 500 µL - 1 mL of lysate) |
Case Study

TRIM67 drives tumorigenesis in oligodendrogliomas through Rho GTPase-dependent membrane blebbing
Journal: Neuro-Oncology
Published: 2023
- Main Technology
- Background
- Methods
- Results
- Conclusions
- Reference
RNA sequencing, total lysate-MS, and co-IP-MS with functional assays including immunofluorescence (IF) staining, and western blotting (WB)
Isocitrate dehydrogenase (IDH) mutant gliomas are grouped into astrocytomas or oligodendrogliomas depending on the codeletion of chromosome arms 1p and 19q. Although the genomic alterations of IDH mutant gliomas have been well described, transcriptional changes unique to either tumor type have not been fully understood. Here, we identify Tripartite Motif Containing 67 (TRIM67), an E3 ubiquitin ligase with essential roles during neuronal development, as an oncogene distinctly upregulated in oligodendrogliomas.
We used several cell lines, including patient-derived oligodendroglioma tumorspheres, to knock down or overexpress TRIM67. We coupled high-throughput assays, including RNA sequencing, total lysate-mass spectrometry (MS), and coimmunoprecipitation (co-IP)-MS with functional assays including immunofluorescence (IF) staining, co-IP, and western blotting (WB) to assess the in vitro phenotype associated with TRIM67. Patient-derived oligodendroglioma tumorspheres were orthotopically implanted in mice to determine the effect of TRIM67 on tumor growth and survival.
TRIM67 overexpression alters the abundance of cytoskeletal proteins and induces membrane bleb formation. TRIM67-associated blebbing was reverted with the nonmuscle class II myosin inhibitor blebbistatin and selective ROCK inhibitor fasudil. NOGO-A/Rho GTPase/ROCK2 signaling is altered upon TRIM67 ectopic expression, pointing to the underlying mechanism for TRIM67-induced blebbing. Phenotypically, TRIM67 expression resulted in higher cell motility and reduced cell adherence. In orthotopic implantation models of patient-derived oligodendrogliomas, TRIM67 accelerated tumor growth, reduced overall survival, and led to increased vimentin expression at the tumor margin.
Taken together, our results demonstrate that upregulated TRIM67 induces blebbing-based rounded cell morphology through Rho GTPase/ROCK-mediated signaling thereby contributing to glioma pathogenesis.
- Demirdizen, Engin, et al. "TRIM67 drives tumorigenesis in oligodendrogliomas through Rho GTPase-dependent membrane blebbing." Neuro-oncology 25.6 (2023): 1031-1043.
FAQs of Co Immunoprecipitation
The difference between IP and Co-IP?
IP, immunoprecipitation, is a method of purification and enrichment of a specific protein by using specific reaction of antigen and antibody.
Co-IP, Co-Immunoprecipitation, is an extension of IP experiment, primarily employed for investigating protein-protein interactions and frequently utilized in the identification of known or unknown constituents within a specific protein complex.
In brief, IP primarily targets the antigen that binds to the antibody, while Co-IP focuses on identifying secondary protein interactions with the primary target proteins rather than antibody.
How to select tag for Co-IP experiments?
The commonly used tags of Co-IP in animal cells include HA, Flag, GFP and MYC, etc., while His and GST tags are often used for prokaryotic cell protein purification. Due to the high histidine background of proteins, His tag is rarely used in mammalian cells.
What are the characteristics of Co-IP/MS service?
1) Investigation of protein-protein interactions and protein complexes;
2) Monitor dynamics of proteins interaction;
3) Study protein-protein interactions in a non-denaturing condition, which is almost physiological;
4) Mapping the interacting domains of proteins.
What is the underlying mechanism that enables protein A to successfully IP protein B when studying protein interactions, while protein B fails to IP protein A?
If A can pull on B, it indicates that there is an interaction between protein A and protein B. However, if B cannot pull on A. At this time, the protein B antibody should be checked first to see if it can IP protein B.
1) If not, it indicates that the B antibody cannot be used for IP, and you can consider changing to another antibody or tagging the protein, using tag antibody to IP B protein;
2) If yes, it may be that B antibody preferentially occupies the binding site of protein B, so that when protein A comes to protein B, protein B no longer exists any binding site for protein A. At this time, it can be considered to change an antibody or add a tag on the protein and use the tag antibody for IP.
Learn about other Q&A of Co-IP.
Publications
Here are some publications from our clients:

- Protein interactors of Spindle Pole Body (SPB) components and septal proteins in fungus Neurospora crassa: A mass spectrometry-based dataset. 2024. https://doi.org/10.1016/j.dib.2023.109980
- Squalene epoxidase induces nonalcoholic steatohepatitis via binding to carbonic anhydrase III and is a therapeutic target. 2021. https://doi.org/10.1053/j.gastro.2021.02.051
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024. https://doi.org/10.1101/2024.07.06.602343
- Hyperfunction of post-synaptic density protein 95 promotes seizure response in early-stage Aβ pathology. 2024. https://doi.org/10.1038/s44319-024-00090-0






