TurboID Service
Discover groundbreaking insights into protein interactions with our TurboID Service. This cutting-edge proximity labeling technology is designed to reveal the hidden networks of protein-protein interactions within live cells, offering unparalleled speed, accuracy, and sensitivity.
Submit Your Request Now
×- What is TurboID
- How It Works
- TurboID Service
- Applications
- Sample Requirements
- FAQs
- Case
What is TurboID?
TurboID was developed as an enhanced version of BioID, a mutant of the prokaryotic Escherichia coli biotin ligase BirA. This enzyme converts biotin into a reactive intermediate that covalently labels proximal proteins with biotin. Utilizing TurboID-mediated proximity labeling, researchers can analyze the interactome of proteins associated with a protein of interest, revealing transient or weak protein-protein interactions within the cellular environment. This approach provides valuable insights into complex biological processes.
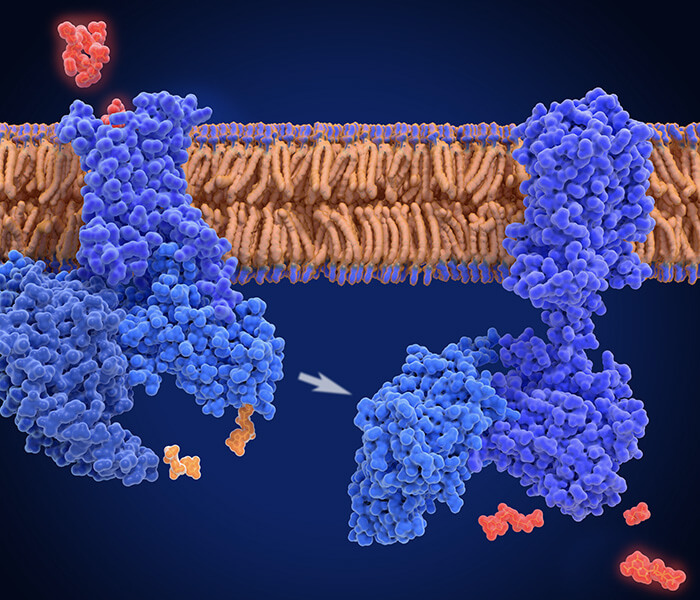
 Figure 1.Yeast display-based selection scheme of TurboID. (Branon, Nature biotechnology, 2018)
Figure 1.Yeast display-based selection scheme of TurboID. (Branon, Nature biotechnology, 2018)
How does TurboID work?
The Principle of Proximity Labeling
Proximity labeling is a recently developed technique for studying protein-protein interactions and subcellular proteomes in live cells. It has been successfully applied in mammalian, bacterial, and plant systems. This approach involves fusing a bait protein with an enzyme possessing specific catalytic activity, which covalently tags nearby endogenous proteins with biotin under enzymatic catalysis. The labeled proteins can then be enriched and analyzed to construct a comprehensive interactome of the bait protein.
By integrating mass spectrometry-based proteomics techniques and high-throughput sequencing, researchers can efficiently identify and characterize biological macromolecules in proximity to the target protein, offering deeper insights into functional interaction networks.
 Figure 2. Proximity labeling technology process. (Ummethum H, Front Genet, 2020)
Figure 2. Proximity labeling technology process. (Ummethum H, Front Genet, 2020)
Principle of TurboID Proximity Labeling
TurboID is an engineered biotin ligase that uses ATP to convert biotin into biotin-AMP, a reactive intermediate capable of covalently labeling proximal proteins. Compared to previous biotin ligase-based proximity labeling methods (such as BioID), TurboID—developed through directed evolution—exhibits significantly higher catalytic efficiency. It reduces labeling time from 18 hours to as little as 10 minutes, allowing higher temporal resolution and expanding its applicability in live-cell and in vivo systems. Biotinylated proteins are subsequently enriched using streptavidin beads and identified through mass spectrometry.
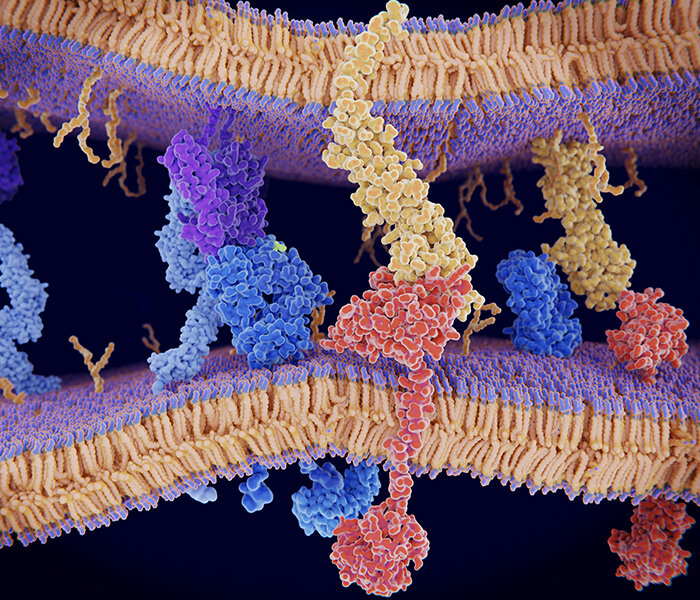
 Figure 3. Proximity-dependent biotinylation catalyzed by TurboID and split-TurboID. (Cho, Nature Protocols, 2020)
Figure 3. Proximity-dependent biotinylation catalyzed by TurboID and split-TurboID. (Cho, Nature Protocols, 2020)
Technical Platforms
Instrument picture from the official website
Our TurboID Service
Our TurboID Service offers researchers a comprehensive platform to explore protein-protein interactions (PPI) with precision and efficiency. Leveraging advanced bioinformatics tools and cutting-edge mass spectrometry technologies, the service supports a wide range of experimental needs, from construct design to data interpretation. This robust service is tailored to provide researchers with high-quality results for understanding complex cellular processes and discovering novel molecular interactions.
We offer the following TurboID Service, but not limited to:
- TurboID Construct Design
- Expression and Biotinylation
- Protein Isolation and Enrichment
- Mass Spectrometry-Based Proteomic Analysis
- Data Interpretation and Reporting
Other optional PPI assay services we provide:
Co-Immunoprecipitation (Co-IP)
IP-MS Protein Interactomics Solution
Proximity Protein Labeling - BioID
Tandem Affinity Purification (TAP)-MS
Learn more about the protein-protein interaction networks
Applications of TurboID Sequence
Mapping Protein-Protein Interactions
Reveals interaction networks by labeling proteins in proximity to a target protein, enabling the identification of novel binding partners and functional pathways.
Investigating Subcellular Localization
Determines protein locations and spatial relationships within cellular compartments.
Studying Protein Dynamics
Analyzes temporal changes in protein interactions during key cellular processes.
Identifying Drug Targets
Uncovers potential therapeutic targets by mapping interactions with specific proteins or complexes.
Our Advantages
- Cutting-Edge Technology: Advanced TurboID proximity labeling for faster and higher-resolution studies.
- Comprehensive Solutions: One-stop service from construct design to high-level data interpretation.
- State-of-the-Art Platforms: Access to premium equipment like Thermo Orbitrap Fusion Lumos and Bruker timsTOF Pro.
- Experienced Team: Expertise in bioinformatics and proteomics ensures reliable results and actionable insights.
- Complete Reporting Process:Include Project completion report, Protein elution silver-stained gel image, Mass spectrometry raw data and analysis results and PPI network diagram
Sample Requirements
| Sample type | Recommended sample size | |
|---|---|---|
| Animal tissues | Hard tissues (bones, hair) | 300-500mg |
| Soft tissues (leaves, flowers of woody plants, herbaceous plants, algae, ferns) | 200mg | |
| Plant tissues | Hard tissues (roots, bark, branches, seeds, etc.) | 3-5g |
| Microbes | Common bacteria, fungal cells (cell pellets) | 100μL |
| cells | Suspension/adherent cultured cells (cell count/pellet) | >1*107 |
| Fluids | Plasma/serum/cerebrospinal fluid (without depletion of high abundance proteins) | 20μL |
| Plasma/serum/cerebrospinal fluid (with depletion of high abundance proteins) | 100μL | |
| Follicular fluid | 200μL | |
| Lymph, synovial fluid, puncture fluid, ascites | 5mL | |
| Others | Saliva/tears/milk | 3-5mL |
| Culture supernatant (serum-free medium cannot be used) | 20mL | |
| Pure protein (best buffer is 8MUrea) | 300μg | |
| FFPE | Each slice: 10µm thickness, 1.5×2cm area | 15-20 slices |
FAQs about TurboID
How does TurboID proximity labeling differ from traditional proximity labeling technologies?
TurboID utilizes a highly efficient yeast enzyme (ApeX2) to catalyze biotinylation with a much faster reaction time (10 minutes to 1 hour) compared to traditional methods like BioID, which often require 18-hour incubation. This results in reduced background noise and higher sensitivity for detecting transient or weak protein interactions.
What are the key advantages of TurboID proximity labeling compared to other high-throughput protein interaction detection methods?
TurboID offers faster labeling, enhanced sensitivity, and reduced background noise. It captures transient or weak protein interactions, works in vivo, and is applicable to low-abundance and membrane proteins. TurboID is versatile, suitable for both cell cultures and animal models, and provides reliable results in physiological conditions.
What types of research applications are suitable for TurboID proximity labeling technology?
TurboID is suitable for applications such as mapping protein-protein interactions, studying protein networks, identifying interaction changes under disease conditions, and examining signal transduction pathways. It is particularly valuable for studying transient or weak interactions, as well as interactions in low-abundance or membrane proteins, both in vitro and in vivo.
How is the protein interaction data generated by TurboID proximity labeling analyzed and interpreted?
Protein interaction data from TurboID labeling is typically analyzed through mass spectrometry to identify labeled proteins. Western blotting and other techniques confirm the interactions. The data is then used to construct protein interaction networks, and bioinformatics tools are employed to interpret the biological significance of these interactions in various contexts.
Learn about other Q&A about other technologies.
TurboID Case study
References
- Branon, Tess C et al. "Efficient proximity labeling in living cells and organisms with TurboID." Nature biotechnology vol. 36,9 (2018): 880-887.
- Ummethum, Henning, and Stephan Hamperl. "Proximity Labeling Techniques to Study Chromatin." Frontiers in genetics vol. 11 450. 12 May. 2020.
- Cho, Kelvin F., et al. "Proximity labeling in mammalian cells with TurboID and split-TurboID." Nature Protocols 15.12 (2020): 3971-3999.