Comprehensive Metabolic Flux Analysis (MFA) Services
At Creative Proteomics, we provide top-tier metabolic flux analysis (MFA) services based on stable isotope labeling (also known as isotope tracing). By incorporating isotope-labeled substrates into metabolic pathways, our approach enables both quantitative and qualitative assessment of metabolic flux. This technique helps determine:
- Metabolic pathway dependencies on specific carbon sources
- Flux distribution at metabolic branch points
- Pathological metabolic shifts in disease models
MFA serves as a powerful tool to analyze metabolic reaction rates, offering deep insights into metabolic network dynamics and their implications in health and disease.
Submit Your Request Now
×- Introduction
- Overview of Services
- Metabolic Pathways
- Advantages
- Platform
- Workflow
- Sample Requirements
- Case Study
- Publication
What Is Metabolic Flux?
Metabolic flux refers to the rate at which metabolites are produced and consumed within a metabolic network under steady-state conditions. It is a critical parameter in cellular physiology, influencing energy production, biosynthesis, and overall metabolic balance.
Fluxomics, the study of metabolic flux dynamics over time, helps researchers:
- Quantify intracellular metabolic changes
- Track metabolic pathway shifts in response to perturbations
- Understand how metabolic imbalances contribute to disease
Metabolic flux can be categorized into:
1. Qualitative Flux Analysis: Determines the direction of metabolite flow using mass isotope distribution (MID), helping identify metabolic shifts (e.g., comparing disease vs. healthy states).
2. Quantitative Flux Analysis: Measures the absolute rate of metabolic conversions (e.g., the amount of substance converted from A to B per cell per hour, in fmol/cell/hr).
Technical Principles of Metabolic Flux Analysis
MFA relies on mass balance equations and the steady-state assumption to quantify metabolic flux. The core methodology involves:
Stable Isotope Labeling (e.g., ¹³C, ¹⁵N)
1. Introduce ¹³C-labeled glucose or ¹³C-labeled glutamine into cell cultures.
2. Labeled carbon atoms get incorporated into downstream metabolites.
3. Different isotopomers (isotope-labeled variants of a metabolite) form based on metabolic pathway activity.
Mass Spectrometry (MS) Analysis
1. High-resolution MS (HRMS) or gas chromatography-MS (GC-MS) detects labeled metabolites.
2. Isotopic distribution patterns reveal pathway activity.
Mathematical Modeling
1. Metabolic flux equations integrate isotopic labeling data.
2. Computational algorithms solve multi-variable equations to estimate metabolic flux rates.
This technique allows precise mapping of metabolic pathways, such as:
- Oxidative vs. reductive TCA cycle flux
- Pentose phosphate pathway (PPP) activity
Amino acid and lipid metabolism shifts
Overview of Metabolomics Flux Analysis Service
At Creative Proteomics, we are dedicated to advancing mass spectrometry-based metabolic flux analysis. Our expertise spans ¹³C-labeled metabolic flux analysis (including the TCA cycle, EMP pathway, and PPP pathway) and ¹⁵N-labeled flux studies (covering organic acids and amino acid metabolism).
Beyond standard metabolomics, we offer cutting-edge metabolic flux detection at both cellular and in vivo (mouse model) levels. Additionally, our customized targeted metabolic flux services provide tailored solutions to help clients overcome technical challenges in metabolic research.
Types of Isotope-Labeled MFA Services
We offer a range of stable isotope tracer metabolic flux analysis options, including:
- ¹³C-labeled glucose MFA (for glycolysis & TCA cycle studies)
- ¹³C-labeled glutamine MFA (for amino acid & anaplerotic flux studies)
- ¹⁵N-labeled glutamine MFA (for nitrogen metabolism research)
- ¹³C dynamic metabolic flux analysis (¹³C-DMFA)
- Isotopic non-stationary metabolic flux analysis (INST-MFA)
Our services are customizable to meet specific research needs. Contact us for tailored experimental designs.
Metabolic Pathways Covered
Our MFA services provide coverage for key metabolic pathways, including:
| Metabolic Pathway | Representative Metabolites |
|---|---|
| Glycolytic pathway | D-Glucose, Glucose 6-phosphate, Fructose 6-phosphate, Fructose-1,6-bisphosphate, Glyceraldehyde 3-phosphate, Dihydroxyacetone phosphate, 3-Phosphoglyceric acid, 2-Phosphoglyceric acid, Phosphoenolpyruvic acid, Pyruvic acid, Lactic acid |
| Pentose phosphate pathway | 6-Phosphogluconic acid, Ribulose-5-phosphate, Ribose-5-phosphate, Xylulose-5-phosphate, Sedoheptulose 7-phosphate, Erythrose-4-phosphate, NADPH |
| Tricarboxylic acid cycle pathway | Citric acid, cis-Aconitic acid, Isocitric acid, 2-Ketoglutaric acid, Succinic acid, Fumaric acid, Malic acid, NADH, FADH₂ |
| Amino acid metabolism | Glycine, Alanine, Serine, Aspartic acid, Glutamine, Glutamic acid, Threonine |
| Fatty acid metabolism | Caprylic acid, Capric acid, Lauric acid, Myristic acid, Myristoleic acid, Palmitic acid, Palmitoleic acid, Stearic acid, Oleic acid, Linoleic acid, Arachidonic acid, Eicosapentaenoic acid (EPA), Docosahexaenoic acid (DHA) |
| One-carbon metabolism | 5,10-Methylene-tetrahydrofolate (THF) (Folate metabolism), S-Adenosylmethionine (SAM) (Methionine cycle), Homocysteine |
| Nucleotide metabolism | ATP, GTP, dTMP, Uracil |
| NADPH and other energy metabolic pathways | NADPH (Pentose phosphate pathway), NADH (TCA cycle), FADH₂ (TCA cycle) |
For additional pathways or specific metabolites of interest, please contact our team.
Advantages of Our Metabolic Flux Analysis Services
- Comprehensive pathway coverage: Includes glycolysis, TCA cycle, amino acid metabolism, and lipid metabolism.
- Advanced mass spectrometry platforms:
- HRMS (Thermo Scientific Q Exactive™) for LC-MS analysis
- GC-MS (Agilent 7890A/5975C) for volatile metabolite analysis
- LC-MS/MS (Agilent 1290-6470A) for targeted quantification
- High-quality data acquisition:
- Extensive quality control (QC) measures ensure accuracy.
- Multi-label isotope tracing (¹³C & ¹⁵N) covering 100+ metabolites.
- Tailored experimental design:
- Metabolic flux analysis at cellular and in vivo levels (mouse models).
- Custom-targeted metabolic flux assays to meet unique research needs.
Instrument Platform
To accommodate various experimental requirements, we utilize different mass spectrometry platforms for analysis.

UHPLC-HRMS: Q Exactive™ (Thermo Scientific Orbitrap MS)
https://www.thermofisher.com

(GC-MS) 7890B/5977A (Agilent GC-MS)
https://www.agilent.com

(UHPLC-MS/MS) 1290-6470A
Workflow of Metabolic Flux Analysis
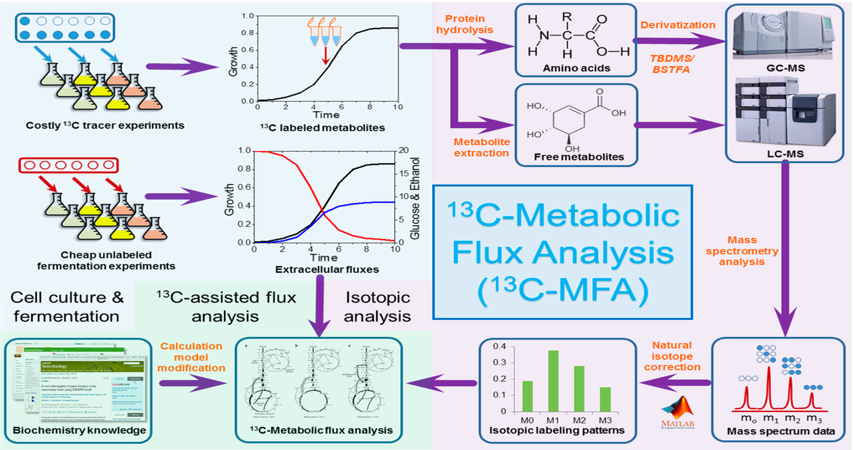 General workflow of steady state ¹³C-Metabolic Flux Analysis (MFA).
General workflow of steady state ¹³C-Metabolic Flux Analysis (MFA).
(https://doi.org/10.3390/bioengineering3010003)
Sample Requirements & Submission Guidelines
| MFA Type | Recommended Biological Replicates | Sample Type | Minimum Sample Size | Labeling Substrates |
|---|---|---|---|---|
| ¹³C-labeled MFA | ≥6 per group | Cells (adherent/suspension) | ≥4×10⁶ cells | ¹³C-glucose, ¹³C-glutamine |
| ¹⁵N-labeled MFA | ≥6 per group | Cells (adherent/suspension) | ≥4×10⁶ cells | ¹⁵N-glutamine |
| Custom MFA | Contact us | Tissue, biofluids | Varies | Custom isotopic tracers |
For customized metabolic flux analysis, please contact our specialists to discuss project requirements.
Challenges in Metabolic Flux Studies & Solutions
Key Challenges
1. Tissue heterogeneity complicates accurate metabolic profiling.
2. Eukaryotic metabolism compartmentalization requires specialized analysis.
3. Lack of single-cell and organelle-specific metabolomics methods.
Solutions
- Cell Sorting Prior to Analysis: Fluorescence-activated cell sorting (FACS) enables cell-type-specific metabolic flux studies.
- Subcellular Fractionation Techniques: Isolation of mitochondria & lysosomes for targeted flux studies.
- Mass Spectrometry Imaging (MSI): Spatial metabolomics at subcellular resolution for in vivo flux analysis.
FAQ
How do I choose the right isotopic tracer for metabolic studies?
Selecting an appropriate tracer depends on the metabolic pathway under investigation.
Radioactive tracers (e.g., ¹⁸F, ³H, ¹⁴C)
- High sensitivity and specificity
- Ideal for tracking predefined pathways
- Require specialized facilities due to radioactivity and half-life constraints
Stable isotope tracers (e.g., ²H, ¹³C, ¹⁵N)
- Lower sensitivity but allow deeper labeling penetration
- Enable simultaneous tracking of multiple pathways
- More suitable for long-term and repeated dosing studies
Among stable isotopes, ¹³C-labeled glucose is the most commonly used tracer for central carbon metabolism due to its efficiency and cost-effectiveness. Other labeled molecules (e.g., ¹⁵N, ²H) are increasingly used to study lipid, ketone body, and amino acid metabolism.
What are the different methods of tracer administration, and how do I choose the best one?
The delivery method should align with the research objective and biological system. Common methods include:
- Oral administration (diet or gavage feeding) – Mimics natural nutrient intake but may result in variable absorption.
- Intravenous (IV) infusion – Provides precise control over tracer levels and minimizes fluctuations but is invasive and costly.
- Intraperitoneal (IP) injection – Allows direct systemic absorption but may introduce variability in uptake rates.
Among these, IV infusion is widely used for in vivo metabolic studies due to its ability to maintain a constant tracer concentration. However, its invasiveness and high cost make it less suitable for dietary metabolism studies.
Regardless of the method, standardizing experimental conditions is crucial to obtaining reproducible and biologically meaningful results.
What are the key challenges in in vivo Metabolic Flux Analysis (MFA)?
In vivo MFA faces several technical limitations compared to in vitro studies:
- Difficulties in measuring uptake and secretion rates – Organ-to-organ metabolite exchange complicates direct rate determination.
- Short labeling durations – In vivo studies often have brief time windows, making it difficult to achieve steady-state labeling required for traditional MFA.
- Limited access to tissue samples – Blood samples are often the only accessible source, restricting tissue-specific metabolic insights.
To address these challenges, researchers are adopting INST-MFA (Isotopically Non-Steady-State MFA), which uses advanced computational models and transient isotope labeling data to infer metabolic fluxes more accurately.
How can I improve the accuracy of in vivo MFA experiments?
To enhance data reliability:
- Choose the right tracer based on the metabolic pathways of interest.
- Optimize delivery methods to maintain consistent isotope labeling.
- Use advanced modeling techniques, such as INST-MFA, to overcome non-steady-state limitations.
- Standardize experimental conditions, including sampling timing and environmental variables, to improve reproducibility.
By integrating these strategies, researchers can achieve more accurate and comprehensive insights into metabolic dynamics in physiological and disease contexts.
Learn about other Q&A.
Case Study: Client Success Stories
YAP mediates compensatory cardiac hypertrophy through aerobic glycolysis in response to pressure overload
https://doi.org/10.1172/JCI150595
- Background
- Client Focus
- Methods
- Results
- Conclusion
Cardiac hypertrophy is the heart's adaptive response to increased workload, such as pressure overload (PO). The Yes-associated protein (YAP), a central player in the Hippo signaling pathway, regulates organ size and cell proliferation. Recent research indicates that YAP facilitates compensatory cardiac hypertrophy by enhancing aerobic glycolysis.
In this study, our team provided U-13C-glucose stable isotope tracing services to investigate how YAP activation influences metabolic pathways in cardiomyocytes under pressure overload conditions. This advanced analysis allowed precise tracking of glucose carbon atoms through glycolytic and related pathways, shedding light on the metabolic reprogramming associated with cardiac hypertrophy.
- Animal Model: Mice underwent transverse aortic constriction (TAC) to induce pressure overload, followed by assessments of cardiac hypertrophy and function.
- Metabolic Flux Analysis: We administered U-13C-glucose to TAC mice and collected cardiac tissues to evaluate 13C incorporation into metabolites. Using an Agilent Technologies system with an Infinity Lab UPLC and a 6545 Q-TOF mass spectrometer, we conducted high-resolution metabolite analysis.
- Metabolomic Profiling: We quantified glycolytic intermediates, tricarboxylic acid (TCA) cycle metabolites, and related pathways to determine the impact of YAP activation on cardiac metabolism.
 (A–D) Time-course analysis of glycolytic flux after PO in freshly isolated AMVMs from control mice.
(A–D) Time-course analysis of glycolytic flux after PO in freshly isolated AMVMs from control mice.
- YAP Activation and Glycolysis: YAP activation led to upregulation of glucose transporter 1 (GLUT1) and increased glycolytic flux, as evidenced by enhanced 13C incorporation into glycolytic intermediates.
- Metabolic Reprogramming: Enhanced glycolysis was associated with increased pyruvate entry into the TCA cycle, supporting the energetic and biosynthetic demands of hypertrophic cardiomyocytes.
- Cardiac Function: Mice with YAP activation exhibited improved cardiac function and adaptive hypertrophy in response to pressure overload, suggesting a protective role of metabolic reprogramming.
This study highlights YAP's crucial role in mediating metabolic adaptations during cardiac hypertrophy. Our U-13C-glucose stable isotope tracing service provided essential insights into the glycolytic enhancements associated with YAP activation, identifying potential therapeutic targets for managing cardiac hypertrophy and preventing heart failure.
Publication

- Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy. van Hummel, A., Taleski, G., Sontag, J. M., Feiten, A. F., Ke, Y. D., Ittner, L. M., & Sontag, E. 2023. https://doi.org/10.1111/nan.12931
- YAP mediates compensatory cardiac hypertrophy through aerobic glycolysis in response to pressure overload. Kashihara, T., Mukai, R., Oka, S. I., Zhai, P., Nakada, Y., Yang, Z., ... & Sadoshima, J. 2022. https://doi.org/10.1172/JCI150595
- Annexin A2 Controls Lipid Composition Necessary for Junctional Stability in Response to Sphingosine 1-Phosphate (Doctoral dissertation). Sveeggen, T. 2022. https://oaktrust.library.tamu.edu/handle/1969.1/198138
- Mechanisms underlying neonate-specific metabolic effects of volatile anesthetics. Stokes, J., Freed, A., et al. 2021. https://doi.org/10.7554/eLife.65400
References
- Antoniewicz, M. R. (2015). Methods and advances in metabolic flux analysis: A mini-review. Journal of Industrial Microbiology & Biotechnology, 42(3), 317-325. 10.1007/s10295-015-1585-x
- Li, M., Wang, Y., Wei, X. et al. AMPK targets PDZD8 to trigger carbon source shift from glucose to glutamine. Cell Res 34, 683–706 (2024). https://doi.org/10.1038/s41422-024-00985-6
- P., A. C., Narushima, H., L., C. R., et al. (2024). Atlas of fetal metabolism during mid-to-late gestation and diabetic pregnancy. Cell, 187(1), 204-215. DOI: 10.1016/j.cell.2023.11.011
- M., J. M., H., J. C., Zhang, Z., et al. (2024). Glycine homeostasis requires reverse SHMT flux. Cell Metabolism, 36(1), 103-115. https://doi.org/10.1016/j.cmet.2023.12.001
- Tran, H. D., Kim, D., Kesavan, R., et al. (2024). De novo and salvage purine synthesis pathways across tissues and tumors. Cell.https://doi.org/10.1016/j.cell.2024.05.011
- Verkerke, P. R. A., Wang, D., Yoshida, N., et al. (2024). BCAA-nitrogen flux in brown fat controls metabolic health independent of thermogenesis. Cell, 187(10), 2359-2374. https://doi.org/10.1016/j.cell.2024.03.030
- Zhou, X., Liu, Y., Shen, Y., et al. (2024). Rescue of cardiac dysfunction during chemotherapy in acute myeloid leukemia by blocking IL-1α. European Heart Journal. https://doi.org/10.1093/eurheartj/ehae188