Protein-Protein Interaction Networks Service
Protein-protein interactions (PPIs) are at the core of virtually every cellular function, influencing key biological processes such as signal transduction, metabolic regulation, and cellular development. Understanding these interactions is critical for exploring the molecular mechanisms behind diseases and advancing therapeutic strategies. At Creative Proteomics, we provide cutting-edge tools to detect, analyze, and map PPIs in both in vivo and in vitro settings.
Submit Your Request Now
×- Featured Protein-Protein Interaction Service
- Case
- Sample Requirements
- Overview
- FAQs
- Publications
Featured Protein-Protein Interaction Service
Creative Proteomics provides protein-protein interaction detection services, including Yeast Two-hybrid, Bio-Layer Interferometry (BLI), Co-Immunoprecipitation (CO-IP), and GST pull-down, which enable qualitative and quantitative analysis of interactions between proteins, small molecules, antibodies, and Fab fragments, among others. Each method has its applicability, and the selection can be based on experimental requirements, including in vivo methods (such as CO-IP and Yeast Two-hybrid) and in vitro methods (such as GST pull-down, Bio-Layer Interferometry, and Surface Plasmon Resonance).
Service Contents
- In vivo
- In vitro
In vivo
Uses antibodies to isolate target protein and its interactors, identified via mass spectrometry.
Identifies protein-protein interactions by immunoprecipitation and mass spectrometry to analyze complexes precisely.
- Fluorescence Resonance Energy Transfer (FRET)
Detects protein interactions by measuring energy transfer between fluorophores within 10 nm distance.
Proximity labeling to identify transient/weak interactions using modifying enzymes and mass spectrometry.
- Yeast Two-hybrid
Detects protein interactions via reconstitution of a transcription factor activating a reporter gene in yeast.
Dual-tag affinity purification improves specificity, followed by mass spectrometry for interaction analysis.
- Bimolecular Fluorescence Complementation (BiFC)
Visualizes interactions by reassembling fluorescent protein fragments fused to interacting partners in living cells.
Combines isotopic labeling with CoIP and MS for quantitative interaction analysis under different conditions.
Uses chemical crosslinkers and MS to map spatial protein interaction sites and relationships.
Employs isotopic labeling to study protein dynamics, quantitative changes, and modifications via MS.
Enhanced version of BioID with faster biotinylation for detecting nearby proteins in live cells.
In vitro
Immobilized GST-tagged proteins capture interactors for in vitro interaction studies.
- Bio-Layer Interferometry (BLI)
Measures biomolecular interactions label-free in real time, quantifying binding kinetics and affinities.
Measures molecular interactions label-free, providing detailed kinetics and mass information.
Detects direct interactions by using labeled bait proteins to probe for targets on membranes.
- Protein Microarray
Immobilized proteins detect interactions with fluorescence, enabling high-throughput functional analysis.
- Phage Display
Links protein-genetic information using bacteriophages to screen for binding partners or peptides.
Identifies drug-protein binding via decreased target sensitivity to protease degradation.
Protein Interaction Case study
Why Creative Proteomics?
- Abundant small molecule database resources for the identification and screening of interacting small molecules.
- Capable of detecting both in vitro and in vivo protein interactions, encompassing weak and strong interactions, as well as transient and stable interactions.
- Custom Assay Development: Custom protocol & assay development as well as assay validation on custom samples
- Data Analysis: Comprehensive report including data analysis and guidance on possible future directions
Sample Requirements
| Sample type | Recommended sample size | |
|---|---|---|
| Animal tissues | Hard tissues (bones, hair) | 300-500mg |
| Soft tissues (leaves, flowers of woody plants, herbaceous plants, algae, ferns) | 200mg | |
| Plant tissues | Hard tissues (roots, bark, branches, seeds, etc.) | 3-5g |
| Microbes | Common bacteria, fungal cells (cell pellets) | 100μL |
| cells | Suspension/adherent cultured cells (cell count/pellet) | >1*107 |
| Fluids | Plasma/serum/cerebrospinal fluid (without depletion of high abundance proteins) | 20μL |
| Plasma/serum/cerebrospinal fluid (with depletion of high abundance proteins) | 100μL | |
| Follicular fluid | 200μL | |
| Lymph, synovial fluid, puncture fluid, ascites | 5mL | |
| Others | Saliva/tears/milk | 3-5mL |
| Culture supernatant (serum-free medium cannot be used) | 20mL | |
| Pure protein (best buffer is 8MUrea) | 300μg | |
| FFPE | Each slice: 10µm thickness, 1.5×2cm area | 15-20 slices |
Overview of Protein-Protein Interactions
Protein-protein interactions (PPIs) are essential regulators of numerous cellular processes. They represent specific physical contacts between proteins that drive critical biochemical events. As key components of the cellular interactome, PPIs are closely linked to disease mechanisms and biological regulation.
By studying PPIs, researchers can uncover the molecular basis of cellular responses and better understand complex biological systems. While some proteins function as monomers, most operate through interactions with other proteins or as part of multi-protein complexes. Therefore, characterizing these interactions is fundamental to understanding protein function and the underlying mechanisms of health and disease.
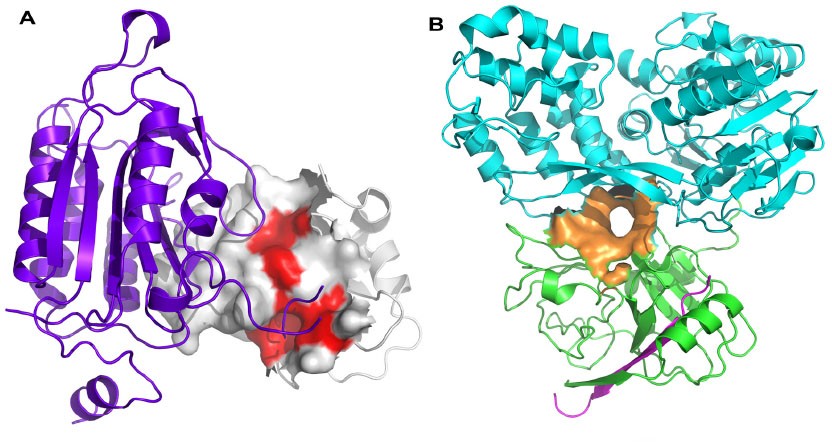 Figure 1. Protein-protein interaction hot spots and allosteric sites. (Turnbull A P. et al.; 2014)
Figure 1. Protein-protein interaction hot spots and allosteric sites. (Turnbull A P. et al.; 2014)
See more about protein-protein interactions
What Is Protein-Protein Interactions In Proteomics?
Enormous molecular processes are regulated via a large number of protein components organized by Protein-Protein Interactions, which refer to intentional physical contacts established between two or more proteins and resulted in specific biochemical events. Such interactions undertake at the core of the entire interactomics system of the living cells, unsurprisingly, specific PPIs are identified with a correlation of multiple diseases.
Protein-protein interactions form the basis of life phenomena, and studying these interactions can elucidate the mechanisms behind biological responses and reveal the essence of life phenomena. While some proteins can exert their functions as monomers, the majority of proteins function through protein-protein interactions or by forming protein complexes. Therefore, investigating protein interactions has become an essential aspect of studying protein function and mechanisms of action.
If we want to study whether one protein interacts with another protein or with several other proteins, the first step is to identify the target protein and then identify other associated proteins. This process ultimately enables us to achieve our experimental objectives.
Protein Protein Interaction Detection Technique
In Vivo Detection Method:
Co-IP
Co-IP combined with Mass Spectrometry (MS), often referred to as Co-IP-MS or IP-MS (Co-Immunoprecipitation Mass Spectrum, Immunoprecipitation Mass Spectrum), is an important tool for discovering and studying protein-protein interactions. Co-IP-MS or IP-MS is a classical method based on the specific interaction between antibodies and antigens, used to investigate protein-protein interactions. It is an effective approach for determining the physiological interactions between two proteins within intact cells.
When cells are lysed under non-denaturing conditions, many protein-protein interactions present inside the cells are preserved. By using specific antibodies or other affinity reagents to bind to the target protein, Co-IP extracts the target protein along with its interacting proteins from complex samples. Subsequently, mass spectrometry is employed to identify the interacting partners of the target protein. SILAC technology combined with Co-IP-MS technology can be used for high-throughput quantitative analysis of protein interactions networks in specific cases. (For a detailed introduction to the Co-Immunoprecipitation experiment, please refer to the Co-IP experimental protocol.)
FRET
Fluorescence Resonance Energy Transfer (FRET) is an effective method used for qualitative and quantitative detection of interactions between biomacromolecules. Depending on the configuration of the fluorescence microscope, FRET can be applied at various levels, such as in solution, cell suspensions, multicellular systems, single cells, cell membranes, and cellular organelles, to study interactions, distances, and kinetic properties between biomacromolecules.
In the field of life sciences, FRET technology is a powerful tool for detecting nanoscale distances and nanoscale distance changes between biomacromolecules in living organisms. It can be used to determine whether two protein molecules directly interact within a single cell. As mentioned earlier, FRET occurs when the emission spectrum of the donor fluorophore overlaps with the absorption spectrum of the acceptor fluorophore, and the distance between the two probes is within the range of 10 nm. In biological systems, if the distance between two protein molecules is within 10 nm, it is generally considered that these two protein molecules have a direct interaction.
TAP- Mass Spectrometry
Regardless of which tag is used as the bait protein to pull down the target protein, it can only serve as a crude enrichment step. When the relationship between the proteins is known or predicted before the experiment, Western Blotting can be used for validation. However, when discovering a new, unknown interaction, a high-throughput, high-resolution technique is required, and this is where mass spectrometry comes into play.
The steps of affinity purification are similar to Co-IP, but single-tag purification efficiency is low, and there may be a higher amount of residual non-specific proteins. Therefore, TAP has been developed, which utilizes dual-tag purification. TAP can use two different tags in combination, but it is essential to note that no tag combination is universally ideal. The choice of tags mainly depends on their characteristics, the properties of the target protein, such as stability and solubility, and the fusion compatibility between the tag and the target protein. The protein complex obtained from affinity purification is then subjected to mass spectrometry technology for identification and analysis.
BioID-MS
Proximity labeling through the use of modifying enzymes is a novel PPI screening method that enables the detection of transient and weak interactions. By fusing the modifying enzyme with the protein of interest, it can label nearby and potential interacting proteins. This type of technique includes BioID, a selective proximity labeling method for the proteome. Compared to traditional protein interaction discovery methods, BioID technology has significant advantages and is suitable for identifying transient, weak interactions, insoluble cellular structures, and dynamic processes within organisms.
TurboID
Compared to BioID, TurboID has higher catalytic efficiency and lower temperature requirements. TurboID can biotinylate neighboring proteins within a range of approximately 10nm in live cells. Finally, the biotinylated proteins are enriched and purified using streptavidin-coupled magnetic beads, followed by mass spectrometry analysis to identify the neighboring proteins of the target protein in the physical space.
In Vitro Detection Method
Glutathione S-Transferase (GST) Pull-down
GST pull-down is a technique similar to Co-IP, but with a difference that GST pull-down is used for studying protein interactions in vitro, while Co-IP is more commonly used for intracellular interactions. In this method, a fusion protein containing a GST-Tag is used to pull down interacting proteins. The fusion protein is then immobilized and precipitated using glutathione resin beads that can specifically bind to the GST tag. Subsequently, SDS-PAGE is used to separate the protein complexes. Similarly, other tags such as Strep, FLAG, MYC, etc., can also be added to the proteins. However, this method cannot reveal the natural interactions that occur between proteins. (Detailed GST pull-down technology introduction)
Far-Western Blotting
Far-Western blotting is a method derived from Western blotting, specifically designed for studying protein-protein interactions. This technique utilizes labeled or antibody-detected "bait" proteins to detect "prey" target proteins on a transfer membrane. If the bait protein interacts with the target protein, specific bands can be detected using the specificity of the bait protein. Far-Western blotting has been employed to study interactions between proteins involved in DNA replication and repair, and it is considered a rapid, efficient, and reproducible method for investigating protein-protein interactions.
This method allows for the detection of direct interactions between two proteins. If the interaction is indirect, further testing can be conducted using candidate proteins. Due to its high sensitivity, Far-Western blotting has found wide applications in various research fields. (For a detailed introduction to Far-Western blotting technique, please refer to the Far-Western blot experimental protocol.)
DARTS
DARTS (Drug Affinity Responsive Target Stability) is a new technique developed based on the reduced sensitivity of target proteins to protease degradation caused by the binding of small molecule drugs to their target proteins. This technique can be used for drug screening and target identification without chemical modification of the compound, as well as not dependent on drug activity.
SPR
Surface plasmon resonance, or SPR, has been used to characterize and measure interactions between biomolecules since it was first applied to the study of processes on metal surfaces and gas sensing in the early 1980s. The number of publications employing SPR biosensors for medical diagnostics, environmental monitoring, food safety, and security-related analyses has grown, as has the development of SPR sensors for detecting chemical and biological species.
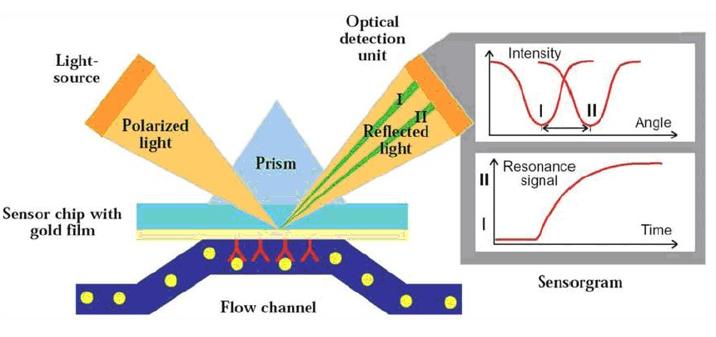 Surface plasmon resonance principle
Surface plasmon resonance principle
This imaging technology provides spatial resolution for protein interactions, allowing detailed studies of proteins using high-throughput microarray techniques. It enables multiplex detection of different proteins without compromising sensitive regions. Fluorescence detection is a widely used imaging technique that allows visualization of proteins labeled with fluorescent dyes. However, label-free techniques are also essential as they eliminate potential impacts of labeling on protein function.
It is crucial to note that unlike label-based methods, label-free techniques directly measure the intrinsic physical properties of proteins, providing additional information such as the mass and size of each protein. Surface Plasmon Resonance Imaging (SPRi) is a label-free in situ detection and molecular binding study technique.
Protein Microarrays
Protein microarrays are chips or slides where various proteins are systematically immobilized on a solid support surface. Samples containing labeled or fluorescently tagged proteins are hybridized with the probe proteins on the microarray. Unbound proteins are washed away, and the fluorescence intensities at each spot on the microarray are measured using a fluorescence scanner or confocal laser scanning technology. By analyzing the fluorescence intensities, protein-protein interactions can be studied, ultimately leading to the determination of protein functions. This method is rapid, simple, and high-throughput, allowing the detection of low abundance and low molecular weight proteins that are often challenging to identify. However, the specificity of binding to the target protein needs improvement.
Phage Display
Phage display technology is a laboratory technique used to study protein-protein, protein-peptide, and protein-DNA interactions. It involves using bacteriophages (viruses that infect bacteria) to link proteins to their genetic information. In this technique, the gene encoding the target protein is inserted into the bacteriophage coat protein gene, allowing the bacteriophage to display the protein on its exterior while containing the gene for the protein inside, thus establishing a link between genotype and phenotype. Other proteins, peptides, or DNA sequences can then be screened against these displayed bacteriophages to detect interactions with the displayed protein. Through this process, large protein libraries can be screened and amplified in a process similar to natural selection, known as in vitro selection.
Protein Interaction Assays FAQs
Why are protein-protein interactions important?
Protein-protein interactions are crucial for the normal functioning of cells and organisms. Interactions between proteins can regulate their activity, stability, and function, thereby influencing various physiological activities and metabolic pathways within cells. Understanding protein-protein interactions helps us comprehend key biological processes such as signal transduction, metabolic regulation, and the cell cycle.
How can protein-protein interactions be detected?
There are various methods to detect protein-protein interactions. Commonly used techniques include co-immunoprecipitation, yeast two-hybrid, GST pull-down, biolayer interferometry (BLI), surface plasmon resonance (SPR), and fluorescence resonance energy transfer (FRET). Each method has its own applicability and limitations, and the choice of method depends on the experimental objectives and the nature of the proteins being studied.
How to choose methods for detecting protein-protein interactions?
Table 1. The most popular methods in PPI studies(For other services, please consult)
| Method | Types of PPIs |
|---|---|
| Co-Immunoprecipitation (co-IP) | Stable or strong |
| SILAC-based CoIP-MS | Stable or strong |
| Pull-Down Assay | Stable or strong |
| Crosslinking Protein Interaction Analysis | Transient or weak |
| Label Transfer Protein Interaction Analysis | Transient or weak |
| Far-Western Blot Analysis | Moderately stable |
What difficulties could arise in the study of protein-protein interactions?
Research on protein-protein interactions may encounter a number of difficulties. It may be challenging to capture and identify certain protein interactions in a stable manner because they are fleeting or weak. Furthermore, selecting the best experimental techniques and technologies to examine certain protein interactions can be challenging and demand a high degree of specificity.
How does research into protein-protein interactions help with disease research and treatment development?
Researching protein-protein interactions can help much with disease and treatment discovery. Understanding these interactions can provide new targets and strategies for medication development and illness diagnosis by illuminating the mechanisms behind the occurrence and progression of disease. Additionally, the creation of specific medications that target particular protein-protein interactions may improve treatment effectiveness and minimize unwanted effects.
Learn about other Q&A about other technologies.
Publications
Here are some publications in proteomics research from our clients:

- Pair bonding and disruption impact lung transcriptome in monogamous Peromyscus californicus. 2023. https://doi.org/10.1186/s12864-023-09873-6
- Morphological and genetic screens reveal mechanisms of BiDAC-induced plasma membrane protein degradation. 2024. https://doi.org/10.21203/rs.3.rs-4438596/v1
- Synergistic Combined-proteomics Guided Mapping strategy identifies mTOR mediated phosphorylation of LARP1 in nutrient responsiveness and dilated cardiomyopathy. 2022. https://doi.org/10.1101/2022.10.13.512080
- Regulation of cardiac ferroptosis in diabetic human heart failure: uncovering molecular pathways and key targets. 2024. https://doi.org/10.1038/s41420-024-02044-w
- Trypanosoma cruzi DNA Polymerase β Is Phosphorylated In Vivo and In Vitro by Protein Kinase C (PKC) and Casein Kinase 2 (CK2). 2022. https://doi.org/10.3390/cells11223693
Reference
- Turnbull A P, Boyd S M, Walse B. Fragment-based drug discovery and protein–protein interactions. Research and Reports in Biochemistry, 2014, 4: 13-26.