What is Phenylalanine Metabolism?
Phenylalanine (Phe), an essential amino acid, is fundamental for protein synthesis in plants. Beyond its role in building proteins, Phe is a precursor to a wide array of secondary metabolites such as phenylpropanoids, flavonoids, lignin, and lignans. These metabolites are essential for key plant processes including reproduction, growth, development, and defense against environmental stresses.
Phe metabolism is a complex process involving multiple enzymes and metabolic pathways. Synthesized in the chloroplast from chorismate, a shikimate pathway precursor, Phe formation involves a three-step enzymatic process: arogenate mutase, prephenate aminotransferase, and arogenate dehydratase. Following this, L-phenylalanine ammonia-lyase (PAL) catalyzes the conversion of Phe to trans-cinnamic acid, initiating the phenylpropanoid pathway which leads to the production of numerous secondary metabolites. Understanding these pathways is crucial for insights into plant physiology, growth, and adaptation.
At Creative Proteomics, our phenylalanine metabolism analysis service equips researchers with the necessary tools and expertise to delve into the complexities of plant Phe metabolism, unlocking its full potential for scientific discovery.
Phenylalanine Metabolism Analysis Service by Creative Proteomics
Phenylalanine Metabolism Metabolite Profiling: In plants, at least 25% of photosynthetic products are stored in phenylpropanoids derived from phenylalanine (Phe). Through customized metabolic profiling solutions, we identify and quantify phenylalanine-derived metabolites with high accuracy and sensitivity. This comprehensive profiling provides a detailed characterization of metabolic intermediates and final products, essential for understanding metabolic fluxes and regulatory pathways.
Phenylalanine Metabolism Pathway Gene Expression Analysis: The biosynthetic pathway of phenylalanine in plants involves a complex regulatory network, and its distribution within plants remains to be studied further. Gene expression frequently shows correlations with metabolite analysis. Our service encompasses gene expression analysis, employing techniques like RNA-Seq to investigate the transcriptional regulation of genes implicated in phenylalanine metabolism.
Phenylalanine Metabolism Enzyme Activity Assays: Verifying the catalytic functions of some key enzymes/restriction enzymes is essential for elucidating the biosynthesis mechanism of phenylalanine and its derivatives. We conduct precise enzyme activity assays to measure key enzymatic reactions involved in phenylalanine metabolism, including L-phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and chalcone synthase (CHS).
Techniques and Instrumentation for Phenylalanine Metabolism Analysis
Liquid Chromatography (LC): At Creative Proteomics, we utilize advanced Liquid Chromatography (LC) techniques to achieve high-resolution separation and quantification of phenylalanine and its derivatives. Our system integrates an Amide chromatography gradient, significantly enhancing the separation conditions for polar substances like amino acids.
Mass Spectrometry (MS): Mass Spectrometry (MS) is a cornerstone of our phenylalanine metabolism analysis services. Utilizing cutting-edge MS technology, we can accurately quantify phenylalanine and its metabolic products, providing comprehensive insights into the metabolic pathways. Our MS systems are designed to handle low-concentration and complex samples with exceptional sensitivity and specificity, ensuring precise detection of even minute quantities of phenylalanine and its metabolites.
LC-MS/MS Method Establishment and Optimization: Our LC-MS/MS services include the establishment and optimization of precise quantification methods using reference standards. We employ Multiple Reaction Monitoring (MRM) and Selected Reaction Monitoring (SRM) techniques to enhance the analysis and quantification of amino acids. These methods are particularly effective for managing complex mixtures and delivering detailed ion information.
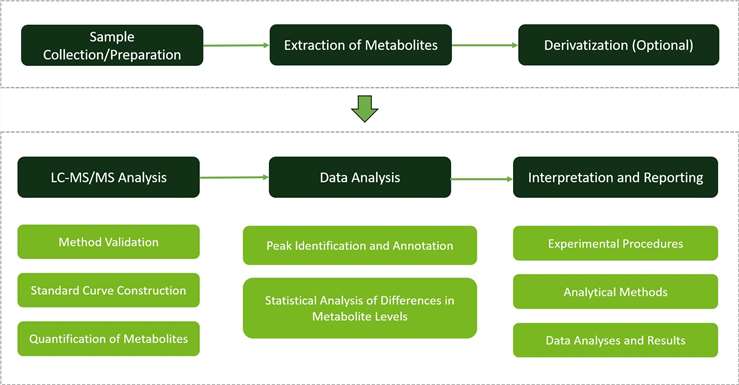
Why Choose Us?
- Expertise and Experience: With over twenty years in plant metabolomics, our team ensures precise analysis of phenylalanine metabolism and associated compounds, guaranteeing reliable results.
- Comprehensive Analysis: Creative Proteomics conducts thorough examinations encompassing enzyme activities and the identification and quantification of metabolites related to phenylalanine metabolism. This approach offers a comprehensive understanding of metabolic pathways, aiding in biosynthetic exploration.
- Advanced Technology: Utilizing cutting-edge instrumentation such as the Thermo Q-Exactive mass spectrometer and Vanquish H UHPLC system, we achieve unparalleled accuracy and sensitivity in analyzing complex samples, particularly those with high impurity levels or low concentrations.
- Customized Solutions: Tailored to individual client needs, our services cater to both research and industrial applications, effectively addressing specific challenges in phenylalanine and metabolite analysis.
- Innovative Approaches: Through pioneering methodologies and ongoing research and development, we continually enhance our analytical capabilities, ensuring we deliver advanced solutions at the forefront of the field.
Applications of Phenylalanine Metabolism Analysis
Secondary Metabolite Research: Understanding the biosynthesis and regulatory mechanisms of secondary metabolites originating from phenylalanine (Phe) is pivotal for enhancing plant traits, including heightened resistance to pests and diseases. Moreover, these metabolites exhibit diverse pharmacological activities, offering promising avenues for drug development and therapeutic treatments.
Agriculture and Crop Improvement: Our services provide new insights into phenylalanine metabolism, which can significantly contribute to developing crops with enhanced growth, yield, and stress tolerance. These advancements are crucial for promoting sustainable agricultural practices and improving crop performance across varied environmental conditions.
Biotechnology Applications: At Creative Proteomics, researchers can utilize metabolomic analysis of phenylalanine pathways to enable the biotechnological production of valuable aromatic compounds, such as flavors and fragrances. This application holds substantial promise in industrial sectors for the efficient production of natural products.
Environmental Interactions: Exploring the roles of phenylalanine-derived compounds in plant-environment interactions enhances researchers' understanding of plant adaptations to changing environmental dynamics. Therefore they can devise more effective strategies to mitigate the impacts of climate change on agricultural systems and ecosystem resilience.
Sample Requirements for Phenylalanine Metabolism Assay
| Sample Type |
Sample Volume |
Notes |
| Blood (Serum/Plasma) |
0.5 - 2 mL |
Collect in a fasting state if possible. Use EDTA or heparin tubes. |
| Urine |
10 - 50 mL |
First morning urine sample preferred. Collect in a sterile container. |
| Tissue Samples |
50 - 100 mg |
Fresh or frozen tissue. Ensure rapid freezing post-collection to -80°C. |
| Cell Culture |
1 - 5 x 10^6 cells |
Harvest cells and wash with PBS. Lyse cells for analysis. |
| Cerebrospinal Fluid (CSF) |
0.2 - 1 mL |
Collect via lumbar puncture, ensure sterility. |
| Saliva |
1 - 2 mL |
Collect unstimulated saliva in sterile tubes. |
| Feces |
5 - 10 g |
Collect in a sterile container. Ensure immediate processing or freezing. |
| Plant Leaves |
100 - 500 mg |
Collect fresh leaves, freeze immediately in liquid nitrogen. |
| Plant Roots |
100 - 500 mg |
Collect fresh roots, clean thoroughly, and freeze immediately. |
| Plant Seeds |
50 - 200 mg |
Collect mature seeds, store in a cool, dry place, or freeze. |
| Plant Stems |
100 - 500 mg |
Collect fresh stems, freeze immediately in liquid nitrogen. |
Each experimental treatment should have more than 6 biological replicates to ensure robust statistical analysis and reliable interpretation of results.
For other sample types not listed above, such as flowers or whole plants, please consult our technical support or sales team for specific requirements and recommendations.
Case. Different Phenylalanine Pathway Responses to Cold Stress Based on Metabolomics and Transcriptomics in Tartary Buckwheat Landraces
Background:
Tartary buckwheat, cultivated for centuries by the Yi people in Southwest China, thrives in challenging environments due to its robust adaptation mechanisms. This crop's diverse landraces are crucial for maintaining biodiversity and meeting local nutritional needs.
Understanding how these landraces respond to cold stress at molecular and metabolic levels is vital for enhancing their resilience and improving agricultural sustainability in mountainous regions.
Samples:
Tartary buckwheat landraces RG and TM, obtained from Yi villages in Liangshan Prefecture, China, and the cultivar "Chuan Qiao NO.2" from the Institute of Crop Science, underwent sterilization with 1% H2O2, followed by germination under controlled conditions (25 °C, 16h light/8h dark).
Cold stress treatment (4 °C, 16h light/8h dark) for 4 days was applied, after which plant materials were promptly frozen in liquid nitrogen and stored at −80 °C. The analysis included 3 replicates for RNA-seq and 6 for metabolomics.
Technical methods procedure:
Total Flavonoids and rutin were extracted with 80% methanol, ultrasonicated, and centrifuged. Supernatants were analyzed using HPLC with a UV detector and a C18 column, using a gradient elution of methanol and acetic acid in water.
Anthocyanins were extracted from seedlings with water/formic acid, ultrasonicated, and centrifuged. The content was analyzed at 520 nm using HPLC with a C18 column and a gradient of water/formic acid and acetonitrile/formic acid.
Total RNA was isolated using Plant RNA Purification Reagent, with quality assessed by a Bioanalyzer and quantified by NanoDrop. cDNA libraries were constructed using the TruSeq RNA Sample Prep Kit. Libraries were sequenced on an Illumina HiSeq xten platform (2 × 150 bp).
Clean reads were processed and aligned to the reference genome using SeqPrep, Sickle, and HIASAT. Assembled reads were analyzed using StringTie, and differential expression was quantified using RSEM and EdgeR. Functional enrichment analysis was performed with Goatools and KOBAS.
Metabolites analysis was performed with a liquid chromatography system coupled to a triple time-of-flight mass spectrometer, using a BEH C18 column and a gradient elution method. PCA and OPLS-DA were used to analyze variations in metabolite profiles. Significance was determined using Student’s t-test with SPSS software, considering P < 0.05 as significant.
Results
Total flavonoid content and specific flavonoids like rutin and cyanidin 3-O-rutinoside were analyzed under cold stress, showing significant increases, particularly in the TM cultivar of Tartary buckwheat.
Genes involved in flavonoid biosynthesis, such as PAL (Phenylalanine Ammonia-Lyase), C4H (Cinnamate-4-Hydroxylase), and F3H (Flavanone 3-Hydroxylase), were significantly upregulated under cold stress in the TM cultivar. Significant differences in gene expression were observed between the cultivars TM, RG, and CQ, indicating varying responses to cold stress.
The combined metabolomics and transcriptomics approach effectively revealed the differential responses of Tartary buckwheat cultivars to cold stress. The TM cultivar exhibited a more robust response, with higher increases in flavonoid content and upregulation of stress-related genes.
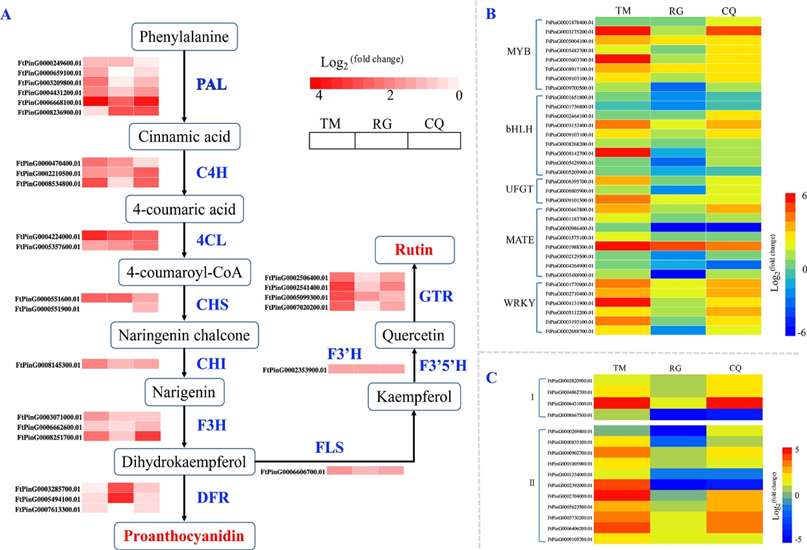 Fig 1. (A) Changes in the expression of genes related to rutin and proanthocyanidin biosynthesis in Tartary buckwheat under cold stress. For the heatmap, each row represents a gene, and three columns represent three Tartary buckwheat genotypes (TM, RG, and CQ). PAL: phenylalanine ammonia lyase; C4H: cinnamate-4-hydroxylase; 4CL: 4-coumarate CoA ligase; CHS: chalcone synthase; CHI: chalcone isomerase; F3H: flavanone-3-hydroxylase; F3′H: flavonoid-3′-hydroxylase; F3′5′H: flavonoid-3′5′-hydroxylase; FLS: flavonol synthase; and DFR: dihydroflavonol reductase. (B) Heatmap of changes in the expression of transcription factors under cold stress. A total of 34 transcription factors (belonging to five groups) were differentially expressed. (C) Heatmap of changes in the expression of genes related to proline and MDA under cold stress. I: genes related to MDA and II: genes related to proline.
Fig 1. (A) Changes in the expression of genes related to rutin and proanthocyanidin biosynthesis in Tartary buckwheat under cold stress. For the heatmap, each row represents a gene, and three columns represent three Tartary buckwheat genotypes (TM, RG, and CQ). PAL: phenylalanine ammonia lyase; C4H: cinnamate-4-hydroxylase; 4CL: 4-coumarate CoA ligase; CHS: chalcone synthase; CHI: chalcone isomerase; F3H: flavanone-3-hydroxylase; F3′H: flavonoid-3′-hydroxylase; F3′5′H: flavonoid-3′5′-hydroxylase; FLS: flavonol synthase; and DFR: dihydroflavonol reductase. (B) Heatmap of changes in the expression of transcription factors under cold stress. A total of 34 transcription factors (belonging to five groups) were differentially expressed. (C) Heatmap of changes in the expression of genes related to proline and MDA under cold stress. I: genes related to MDA and II: genes related to proline.
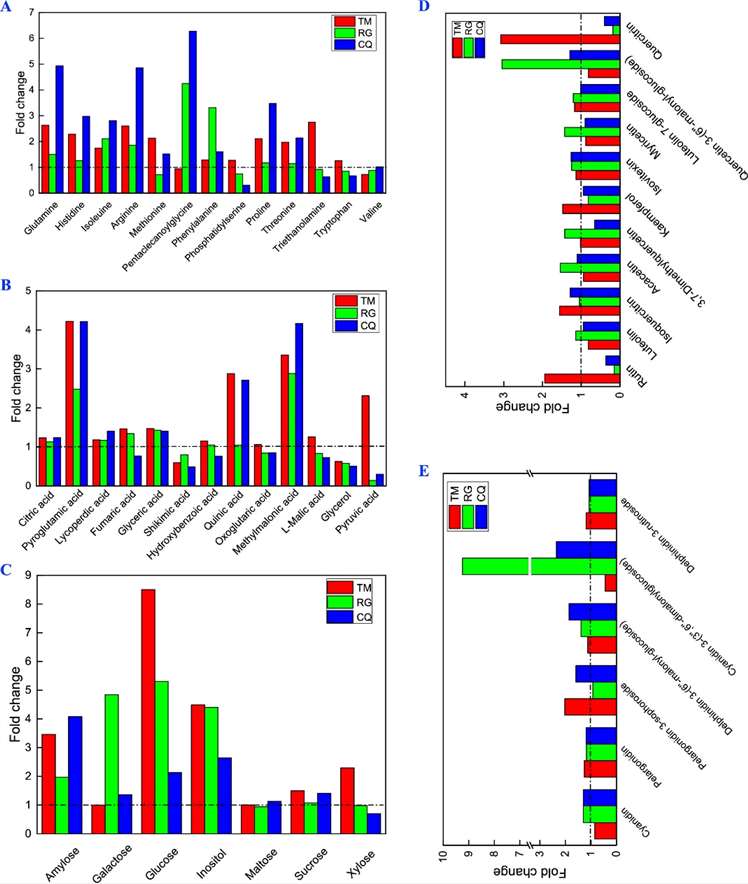 Fig 2. Changes in metabolites in three Tartary buckwheat phenotypes in response to cold stress. (A) Amino acids and their derivatives, (B) organic acid and their derivatives, (C) sugars and their derivatives, (D) flavone biosynthesis, and (E) anthocyanin biosynthesis.
Fig 2. Changes in metabolites in three Tartary buckwheat phenotypes in response to cold stress. (A) Amino acids and their derivatives, (B) organic acid and their derivatives, (C) sugars and their derivatives, (D) flavone biosynthesis, and (E) anthocyanin biosynthesis.
Reference
- Song, Y. (2022). "Different Phenylalanine Pathway Responses to Cold Stress Based on Metabolomics and Transcriptomics in Tartary Buckwheat Landraces." Journal of Agricultural and Food Chemistry 70(2), 687-698.



 Fig 1. (A) Changes in the expression of genes related to rutin and proanthocyanidin biosynthesis in Tartary buckwheat under cold stress. For the heatmap, each row represents a gene, and three columns represent three Tartary buckwheat genotypes (TM, RG, and CQ). PAL: phenylalanine ammonia lyase; C4H: cinnamate-4-hydroxylase; 4CL: 4-coumarate CoA ligase; CHS: chalcone synthase; CHI: chalcone isomerase; F3H: flavanone-3-hydroxylase; F3′H: flavonoid-3′-hydroxylase; F3′5′H: flavonoid-3′5′-hydroxylase; FLS: flavonol synthase; and DFR: dihydroflavonol reductase. (B) Heatmap of changes in the expression of transcription factors under cold stress. A total of 34 transcription factors (belonging to five groups) were differentially expressed. (C) Heatmap of changes in the expression of genes related to proline and MDA under cold stress. I: genes related to MDA and II: genes related to proline.
Fig 1. (A) Changes in the expression of genes related to rutin and proanthocyanidin biosynthesis in Tartary buckwheat under cold stress. For the heatmap, each row represents a gene, and three columns represent three Tartary buckwheat genotypes (TM, RG, and CQ). PAL: phenylalanine ammonia lyase; C4H: cinnamate-4-hydroxylase; 4CL: 4-coumarate CoA ligase; CHS: chalcone synthase; CHI: chalcone isomerase; F3H: flavanone-3-hydroxylase; F3′H: flavonoid-3′-hydroxylase; F3′5′H: flavonoid-3′5′-hydroxylase; FLS: flavonol synthase; and DFR: dihydroflavonol reductase. (B) Heatmap of changes in the expression of transcription factors under cold stress. A total of 34 transcription factors (belonging to five groups) were differentially expressed. (C) Heatmap of changes in the expression of genes related to proline and MDA under cold stress. I: genes related to MDA and II: genes related to proline. Fig 2. Changes in metabolites in three Tartary buckwheat phenotypes in response to cold stress. (A) Amino acids and their derivatives, (B) organic acid and their derivatives, (C) sugars and their derivatives, (D) flavone biosynthesis, and (E) anthocyanin biosynthesis.
Fig 2. Changes in metabolites in three Tartary buckwheat phenotypes in response to cold stress. (A) Amino acids and their derivatives, (B) organic acid and their derivatives, (C) sugars and their derivatives, (D) flavone biosynthesis, and (E) anthocyanin biosynthesis.