Stable isotope, specifically 13C-based metabolic flux analysis, is a common method for accurately quantifying the integrated response of metabolic networks. When a labeled substrate such as [1,2-13C] glucose is metabolized by cells, enzymatic reactions rearrange carbon atoms resulting in specific labeling patterns in downstream metabolites that can be measured with analytical techniques such as mass spectrometry (MS), or nuclear magnetic resonance. Therefore, designing an optimal tracer experiment is an important step in the workflow of high-resolution 13C-MFA, and the substance content of labeling experiments and accuracy of estimating flux rates depend at a large level on the choice of tracer, which, if not properly selected, can directly limit the ability to accurately resolve metabolic flux. For a typical prokaryotic metabolic network, the use of 13C glucose tracers in parallel experiments followed by a detailed analysis of the combined data can provide the most comprehensive metabolic flux data. At Creative Proteomics, our expert teams have designed and employed a protocol based on 13C labeling of microorganisms grown on glucose and subsequent gas chromatography mass spectrometry detection of 13C patterns in protein-bound amino acids.
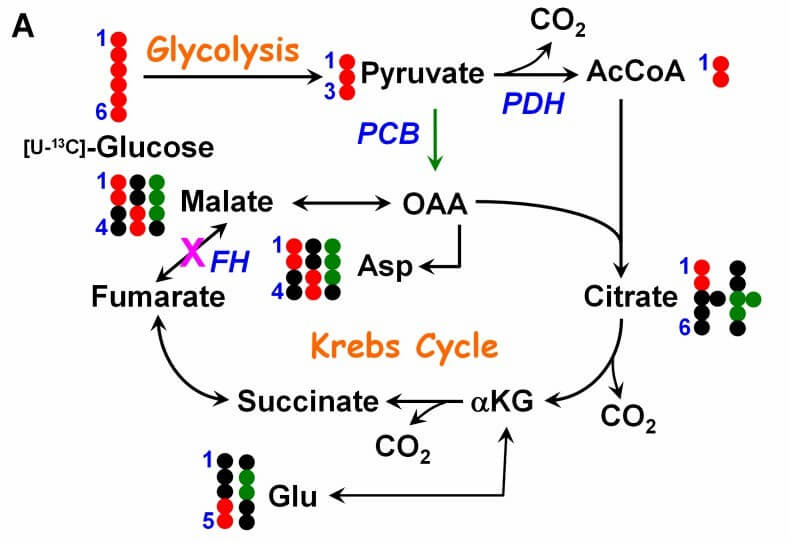 Figure 1. Panel A illustrates the fate of glucose 13C atoms in various glycolytic and Krebs cycle metabolites. (Youfeng, Y.; et al. 2013)
Figure 1. Panel A illustrates the fate of glucose 13C atoms in various glycolytic and Krebs cycle metabolites. (Youfeng, Y.; et al. 2013)
Our Services
Creative Proteomics has developed a novel metabolic flux analysis platform to provide 13C-labeled glucose for 13C-MFA service in a competitive fashion. We can offer a wide range of services to support all research and development activities. By employing 13C uniform labeling of glucose and detecting the 13C-NMR signal intensity of the product, the proportion of the product generated by glucose metabolism is conveniently calculated, so as to infer the metabolic mechanism of microorganisms on multi-carbon sources. Here is our 13C-labeled glucose based metabolic flux analysis procedure:
- 13C-labeled glucose undergoes a glycolytic reaction to produce pyruvate, the final product of the glycolytic reaction. Pyruvate can then undergo pyruvate dehydrogenase to produce acetyl coenzyme A, which enters the tricarboxylic acid cycle (TCA cycle). Glucose, as a substrate for pentose phosphate pathway and serine biosynthesis, can label intermediates in both metabolic pathways. We can obtain the metabolic flow during a certain period of time by analyzing the downstream product labeling of a specific pathway
- After the metabolism of 13C-labeled glucose to pyruvate by glycolysis, it can enter the TCA cycle by both pyruvate dehydrogenase and pyruvate carboxylase, resulting in M+2 and M+3 TCA intermediates, which can be analyzed to obtain information about the different pathways of metabolic flux mediated by the two enzymes
![Panel A shows the atom-resolved fate of 50% [1,2-13C]-glucose through glycolysis and the pentose phosphate pathway.](upload/image/13C-labeled-glucose-for-13C-MFA-2.jpg) Figure 2. Panel A shows the atom-resolved fate of 50% [1,2-13C]-glucose through glycolysis and the pentose phosphate pathway. (Youfeng, Y.; et al. 2013)
Figure 2. Panel A shows the atom-resolved fate of 50% [1,2-13C]-glucose through glycolysis and the pentose phosphate pathway. (Youfeng, Y.; et al. 2013)
Features of Our MFA Platform
- Developed based on the most updated knowledge of biology, bioinformatics and software development
- Widely applicable to a wide range of metabolic system
- Professional bioinformatics teams & personalized bioinformatics analysis services.
- Advanced instrument platform
- Integrated quantitative methodologies and comprehensive solutions for metabolomics
Based on high-performance quantitative techniques and advanced equipment, Creative Proteomics has constantly updating our metabolic flux analysis platform and is committed to offering professional, rapid and high-quality services of 13C-labeled glucose for 13C-MFA at competitive prices for global customers. Our personalized and comprehensive services can satisfy any innovative scientific study demands, please contact our specialists to discuss your specific needs. We are looking forward to cooperating with you!
Reference
- Youfeng, Y.; et al. Metabolic Reprogramming for Producing Energy and Reducing Power in Fumarate Hydratase Null Cells from Hereditary Leiomyomatosis Renal Cell Carcinoma. PLOS ONE. 2013. 8(8): e72179.