Integrated fluxomics and metabolomics analysis is one of the family of "omic sciences". Since metabolites are the final downstream products of genomic, transcriptomic and/or proteomic perturbations, their measurement brings key insights to systems biology. Creative Proteomics designs to expand the availability of metabolomics services to researchers and professional in the life sciences. With advanced metabolic flux analysis (MFA) platform and experienced bioinformatics analysis experts, we are proud to offer integrated fluxomics and metabolomics analysis service to our global customers. Our team of experts can help researchers and professionals identify biomarkers of a pathological state, classify sample groups based on the origin of metabolites, better characterize and understand biological mechanisms.
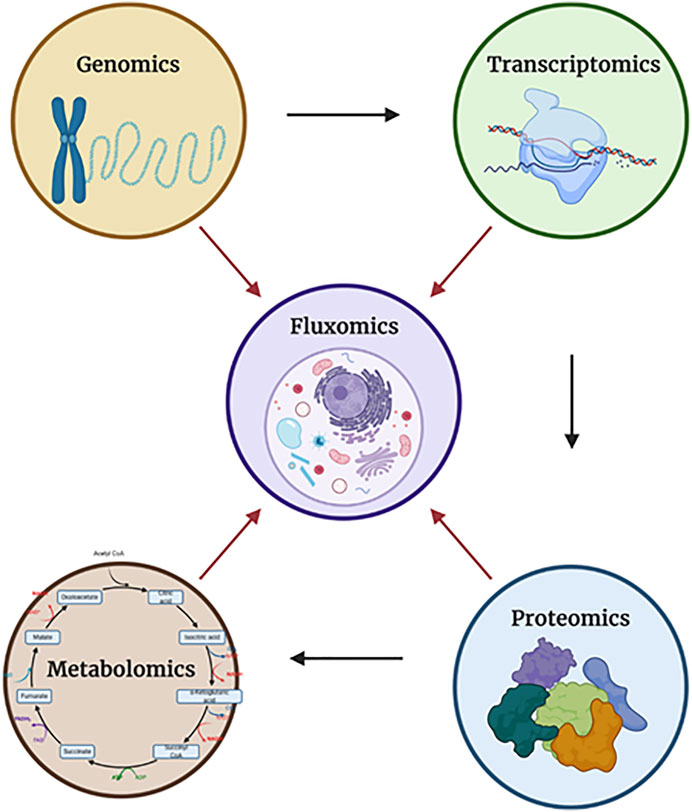 Fig. 1 The relationships between each of the "-omics". (Emwas, Abdul-Hamid, et al. 2022)
Fig. 1 The relationships between each of the "-omics". (Emwas, Abdul-Hamid, et al. 2022)
Metabolomics and fluxomics
Metabolomics and fluxomics methods both focus on small molecules (molecular weight is typically lower than 1000 Da). Metabolomics aims at the identification and quantification of as many metabolites as possible in cells, biofluids and tissues from plant, animal and other biological systems. Fluxomics deals with metabolic fluxes typically based on isotopically labeled compounds (used as tracers), namely, the rate of metabolic conversions in these systems. Fluxomics has become one of the most relevant methods for studying metabolic phenotypes. This method collects data from multiple different -omics fields and presents a full picture of molecular interactions.
Towards a better understanding of metabolite signals and network
Creative Proteomics offers advanced analytical software for processing, characterizing and resolving complex metabolomics data generated by technologies such as MS and NMR. Our comprehensive software platform not only supports the results of all these technologies but also enables the integration of metabolomics and metabolic flux data information for faster and deeper study of biological questions.
In addition, based on liquid chromatography/mass spectrometry (LC-MS), gas chromatography/mass spectrometry (GC-MS) and nuclear magnetic resonance (NMR), we can provide targeted and untargeted metabolomics analysis services for a variety of samples, combined with customizable bioinformatics analysis methods, providing you with one-stop integrated fluxomics and metabolomics analysis services from experimental design, sample detection, to data analysis, which can meet a variety of project needs.
Why choose us?
- Multiple quality control systems for more accurate results
- Rich experience in design and analysis of integrated metabolic flux and omics analysis
- Packaged/customized integrated metabolic flux and omics analysis
- Experienced in large-scale projects
The level of metabolic fluxes is determined by the complex interactions of cellular components. Combining MFA with other omics analysis is a promising strategy for studying the regulatory mechanisms of metabolic systems. Creative Proteomics is a leading service provider of metabolomics services. Our team of experts combine expertise of biologists and analytical chemists, and can provide a tailored design of the biological analysis solution for our clients. If you are interested in our service, please don't hesitate to contact us.
References
- Cocuron, Jean-Christophe, et al. "A combined metabolomics and fluxomics analysis identifies steps limiting oil synthesis in maize embryos." Plant physiology 181.3 (2019): 961-975.
- Babele, Piyoosh Kumar, and Jamey D. Young. "Applications of stable isotope‐based metabolomics and fluxomics toward synthetic biology of cyanobacteria." Wiley Interdisciplinary Reviews: Systems Biology and Medicine 12.3 (2020): e1472.
- Emwas, Abdul-Hamid, et al. "Fluxomics-new metabolomics approaches to monitor metabolic pathways." Frontiers in Pharmacology 13 (2022): 299.