Visualization of metabolic dynamics is important for all types of metabolic studies, including studies on optimization of biological production processes and studies of metabolism-related diseases. Scientists therefore have used PCA score plots to visualize the metabolic distances, and the PCA score plots for this dataset represent a metabolic map that can be used to study the metabolic flow of pathways and organisms for analysis. Principal component analysis is one of the most frequently applied statistical methods in systems biology, and it is used to reduce the dimensionality of the data while retaining most of the variation in the dataset. This reduction is accomplished by identifying linear combinations of variables, called the principal components, that maximally explain the variation in the data. By comparing these principal components to other features, each sample can be represented by a relatively small number of variables. Nowadays, PCA has been widely applied in transcriptomics and fluxomics, where principal components identify linear combinations of gene or enzyme reactions whose variation in activity explains the largest fraction of variability in the set of samples analyzed. Creative Proteomics has developed a new method for analyzing metabolic turnover analysis data by using PCA strategy.
The main goals of PCA
- Determine which parts of the metabolism retain the major variability in flux data
- Relate them to the sample, i.e. the behavior of the organism under specific experimental conditions
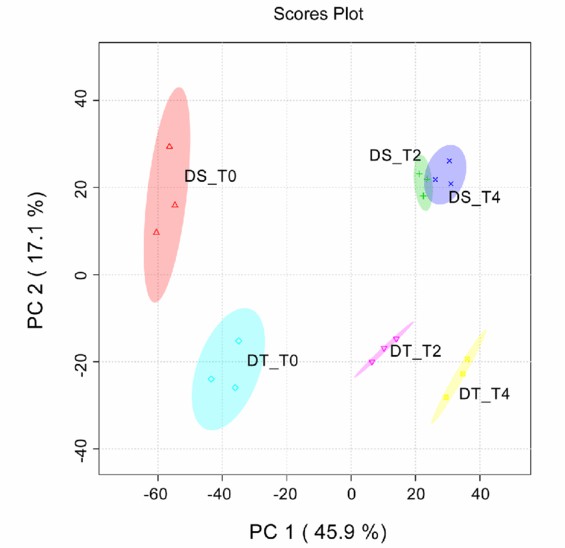 Figure 1. Principal component analysis (PCA) of metabolic profiles of DT and DS under control and drought stress. (You, J.; et al. 2019)
Figure 1. Principal component analysis (PCA) of metabolic profiles of DT and DS under control and drought stress. (You, J.; et al. 2019)
Our Services
Creative Proteomics has developed a novel metabolic flux analysis platform to provide principal component analysis (PCA) service in a competitive fashion. We can offer a wide range of services to support all research and development activities.
- The H NMR spectra are analyzed using principal component analysis to identify metabolites responsible for gender differences
- Various metabolites are identified that are responsible for the observed separations
- Our approach is capable of identifying the metabolites that are important for the discrimination of classes of individuals of similar physiological conditions
Our PCA Methodology
We extract the metabolite ion peaks, and normalize the peaks obtained from multiple experimental samples and QC samples. Then, our expert teams perform a comprehensive analysis via PCA. Here are the details of our analysis process:
- All the metabolites originally identified are regrouped linearly to form a new set of composite variables
- 2-3 composite variables are selected according to the problem being analyzed, so that they reflect as much information as possible about the original variables, thus achieving dimensionality reduction
- Through observing the overall distribution trend among all groups of samples, we can identify possible discrete samples and decide whether to remove the discrete points by considering various factors (number of samples, preciousness of samples, degree of dispersion)
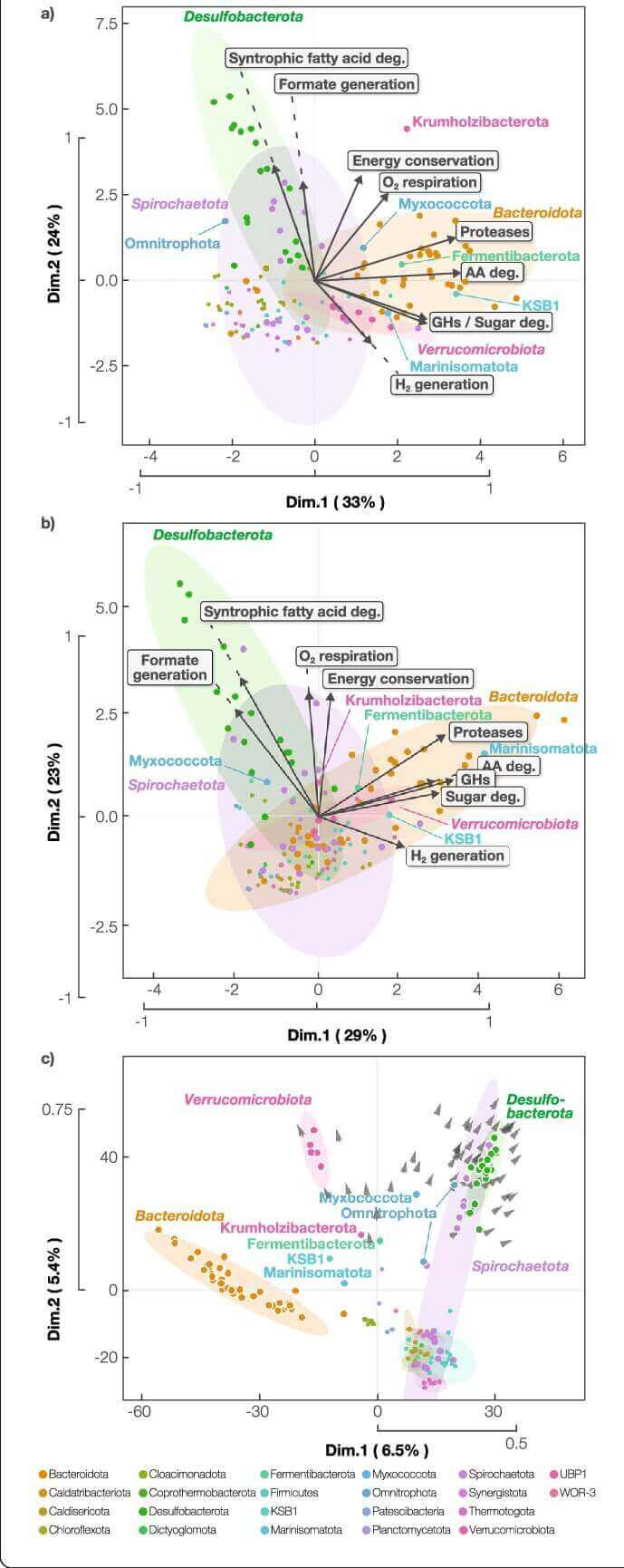 Figure 2. Principal component analysis (PCA) of a metabolic capacities. (Nobu, M, K.; et al. 2020)
Figure 2. Principal component analysis (PCA) of a metabolic capacities. (Nobu, M, K.; et al. 2020)
Features of Our MFA Platform
- Developed based on the most updated knowledge of biology, bioinformatics and software development
- Widely applicable to a wide range of metabolic system
- Professional bioinformatics teams & personalized bioinformatics analysis services.
- Advanced instrument platform
- Integrated quantitative methodologies and comprehensive solutions for metabolomics
Based on high-performance quantitative techniques and advanced equipment, Creative Proteomics has constantly updating our metabolic flux analysis platform and is committed to offering professional, rapid and high-quality services of principal component analysis (PCA) at competitive prices for global customers. Our personalized and comprehensive services can satisfy any innovative scientific study demands, please contact our specialists to discuss your specific needs. We are looking forward to cooperating with you!
References
- You, J.; et al. Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biology. 2019. 19(1).
- Nobu, M, K.; et al. Catabolism and interactions of uncultured organisms shaped by eco-thermodynamics in methanogenic bioprocesses. Microbiome. 2020. 8: 111.