Metabolic flux analysis (MFA) is a core methodology of fluxomics, which enables measures of the rates of all intracellular fluxes in the central metabolism of biological systems. Isotope labeling experiments help MFA by providing information on intracellular fluxes, especially through parallel and circular pathways. During these experiments, mass spectrometry (MS) and nuclear magnetic resonance (NMR) are the main methods to measure the abundances of the isotopomers. Here, Creative Proteomics is proud to offer NMR-based metabolic flux analysis. We are committed to expanding the availability of metabolic flux analysis services to investigators and professionals in a variety of fields while advancing the state-of-the-art in metabolomics research.
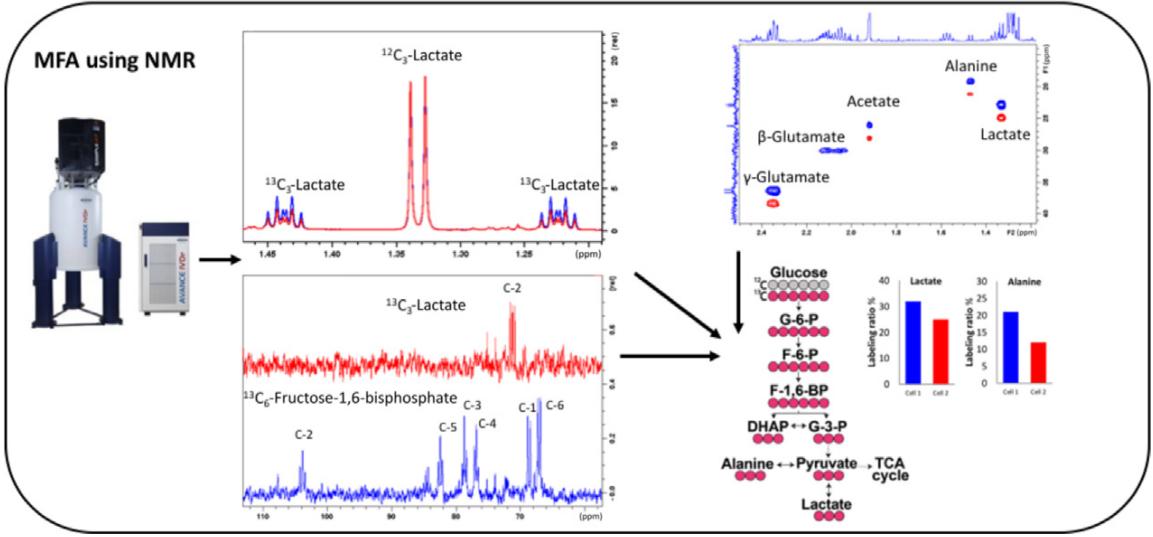 Fig. 1 MFA using NMR. (Radenkovic, Silvia, Ivan Vuckovic, and Ian R. Lanza., 2020)
Fig. 1 MFA using NMR. (Radenkovic, Silvia, Ivan Vuckovic, and Ian R. Lanza., 2020)
NMR—a very powerful tool for metabolic research
Over the past few decades, advances in NMR have propelled more comprehensive metabolite detection and quantification, making it a very powerful tool for metabolic research. NMR offers many unique advantages in metabolic studies, particularly in terms of rigorous and versatile structure elucidation, isotope-filtered selection of molecules, and analysis of positional isotopomer distribution in complex mixtures provided by multinuclear and multidimensional experiments. In addition, NMR allows spatially selective in vivo imaging and dynamic analysis of tissue metabolism in living organisms. Notably, in combination with the use of stable isotope tracers, NMR is a method of choice for exploring the kinetics and delineation of metabolic pathways and networks.
Advantages of NMR-based metabolic flux analysis
- Non-destructive and non-biased
- Relatively high specificity
- Robust, easily quantifiable and reproducible
- Provides information on site-specific label incorporation
- Minimal sample preparation requirements
- Capable of both in vitro and in vivo metabolic flux analysis
Service offering
The pathway and network knowledge generated by NMR-based isotopomer analysis is valuable in guiding the functional analysis of other 'omics' data to enable an understanding of the regulation of biochemical systems. Creative Proteomics is proud to provide NMR-based metabolic flux analysis to help accelerate your research. The general process of analysis services is as follows:
- Experimental design
- Labeling experiment
- NMR sample preparation
- Spectral acquisition of 2-D [13C, 1H] correlation maps
- Spectral processing and flux result
Bioinformatics analysis contents

- Metabolic pathways analysis
- Flux ratio analysis
- Principal component analysis (PCA)
- Hierarchical cluster analysis (HCA)
- KEGG enrichment analysis
- Bi-plot analysis
- Flux balance analysis (FBA)
Why choose us?
- We accept a wide range of biological materials including, but not limited to, cells, tissues, organs, and biofluids
- A comprehensive platform with advanced instrumentation and data analysis software
- Provide a complete analysis report including method interpretation, experimental parameters, data and results analysis
- A team of experts with years of experience in metabolomics, bioinformatics, and statistics
MFA uses MS or NMR to monitor the fate of isotope tracers (such as 13C-glucose and 15N-glutamine), helping researchers to assess the contribution of specific metabolic pathways to the prevailing levels of specific metabolites. Creative Proteomics is a leading provider of metabolic flux analysis services. Our reliable and personalized service can accommodate any innovative research project. Please contact our experts to discuss your specific needs.
References
- Fan, Teresa W-M., and Andrew N. Lane. "Applications of NMR spectroscopy to systems biochemistry." Progress in nuclear magnetic resonance spectroscopy 92 (2016): 18-53.
- Radenkovic, Silvia, Ivan Vuckovic, and Ian R. Lanza. "Metabolic flux analysis: moving beyond static metabolomics." Trends in biochemical sciences 45.6 (2020): 545-546.