Stable isotope distribution in major metabolites helps to study the metabolic distribution of complete organisms. In general, the determination of the isotopic distribution of metabolites in a metabolic network can be used to calculate dynamic patterns such as metabolite synthesis and degradation. At Creative Proteomics, stable-isotope tracing experiments are commonly used to track the flow of stable isotopes in metabolites, and therefore determine the isotopic distribution of metabolites.
Advantage of Our Technology
- High tracking coverage, with the ability to track thousands of metabolites simultaneously with stable isotope labeling
- Highly accurate and reproducible quantification of stable isotope labeled metabolites
- Low false positives for large scale data analysis
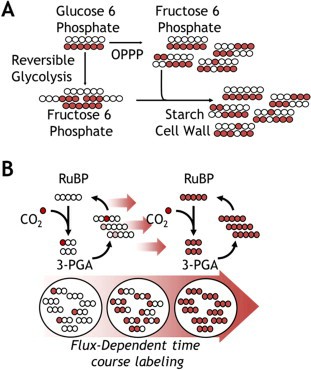 Figure 1. Isotope distribution in heterotrophic and autotrophic pathways. (Allen, D. K.; et al. 2015)
Figure 1. Isotope distribution in heterotrophic and autotrophic pathways. (Allen, D. K.; et al. 2015)
Our Services
Creative Proteomics has developed a novel metabolic flux analysis platform to provide isotopic distribution of metabolites service in a competitive fashion. We can offer a wide range of services to support all research and development activities.
- Creative Proteomics has introduced a new method for efficiently calculating the exact masses in an isotopic distribution by using a dynamic programming approach, and the resolution allows very fine mass structures in isotopic distributions to be seen, even for large molecules
- In addition, our experts establish a strategy for qualitative prediction of intramolecular isotope distribution. Among them are fatty acids, monoterpenes, hopanoids, branched-chain amino acids, amino acids of the trisaccharide family, metabolites synthesized by the shikimate pathway, metabolites of the Krebs cycle, amino acids of the ketoglutaric family, and oxaloacetate. The method has been tested by comparing the natural distribution of carbon with the theoretically predicted isotopic distribution
Process of Isotopic Distribution of Metabolites
- Determination of the exact mass of a compound
- Database search
- Primary identification
- Screening ions with the same molecular weight to determine the mass
- Screening ions with the same molecular weight and elemental composition by isotope distribution comparison to determine the isotope of the compound
- Secondary identification
Screening ions with similar molecular weight, elemental composition and structure by matching the library of standard spectra or theoretical secondary spectra with the actual secondary spectra to determine the tautomer of the compound
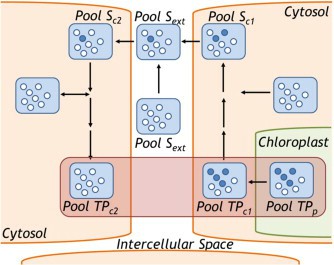 Figure 2. Cellular heterogeneity contributes to spatial labeling differences in metabolites. (Allen, D. K.; et al. 2015)
Figure 2. Cellular heterogeneity contributes to spatial labeling differences in metabolites. (Allen, D. K.; et al. 2015)
Features of Our MFA Platform
- Developed based on the most updated knowledge of biology, bioinformatics and software development
- Widely applicable to a wide range of metabolic system
- Professional bioinformatics teams & personalized bioinformatics analysis services.
- Advanced instrument platform
- Integrated quantitative methodologies and comprehensive solutions for metabolomics
Based on high-performance quantitative techniques and advanced equipment, Creative Proteomics has constantly updating our metabolic flux analysis platform and is committed to offering professional, rapid and high-quality services of isotopic distribution of metabolites at competitive prices for global customers. Our personalized and comprehensive services can satisfy any innovative scientific study demands, please contact our specialists to discuss your specific needs. We are looking forward to cooperating with you!
Reference
- Allen, D. K.; et al. Tracking the metabolic pulse of plant lipid production with isotopic labeling and flux analyses: Past, present and future. Progress in Lipid Research. 2015. 58: 97-120.