Metabolism cannot be studied thoroughly by measuring metabolite concentrations by metabolomics. Equally important is understanding the activity of pathways, so scientists quantify metabolic pathways in terms of the flow of material per unit of time, i.e., metabolic flux. Metabolic pathways are enzyme-mediated biochemical reactions that lead to biosynthesis (anabolism) or breakdown (catabolism) of natural product small molecules within a cell or tissue. Metabolic pathway analysis, a methodology used in metabolic pathway modeling, is widely used to assess inherent network properties and identify meaningful structural and functional units in the metabolic networks.
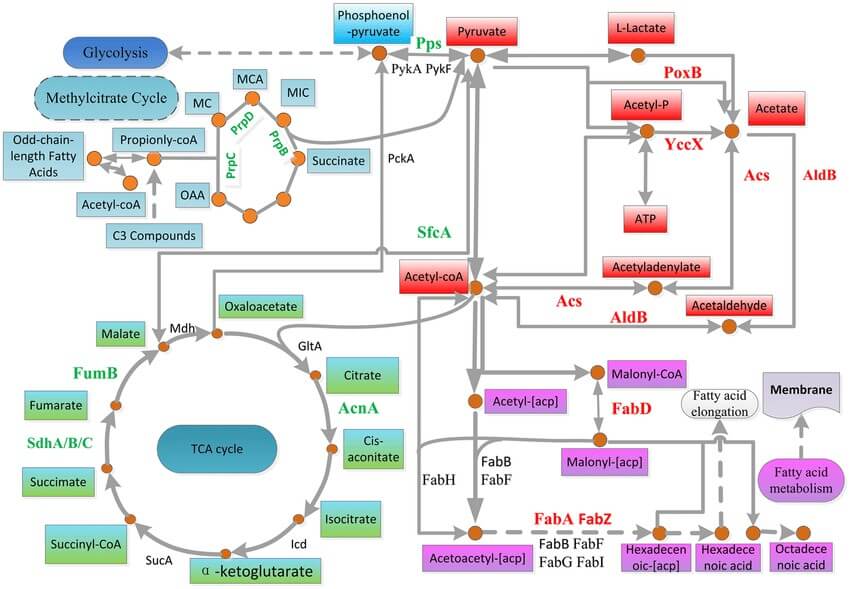 Figure 1. Metabolic pathways potentially associated with Mn oxidation and the Mn(II) stress response. (Zhiyong, W.; et al. 2017)
Figure 1. Metabolic pathways potentially associated with Mn oxidation and the Mn(II) stress response. (Zhiyong, W.; et al. 2017)
Our Services
Creative Proteomics has developed a novel metabolic flux analysis platform to provide metabolic pathways study service in a competitive fashion. We can offer a wide range of services to support all research and development activities.
Identification of metabolic pathway
In metabolomics, identification of metabolic pathways is essential to reveal the underlying biological mechanisms. With the aim of facilitate biological interpretation, our teams have already developed a metabolic pathway identification approach that incorporates all pathways into a global model. And our method is able to identify significant metabolic pathways that contribute to the biological interpretation of metabolomics data.
- Our procedure
- Firstly, the detected metabolites are assigned to pathway blocks based on their metabolic roles as defined by the KEGG pathway database
- Then, the metabolite intensity or concentration data matrices are reconstructed as data blocks based on the metabolite subsets
- Finally, a model is built on these data blocks for assessing the significance of each metabolic pathway for group separation
Discovery of new pathway
Discovery of new metabolic pathways can be achieved by tracking the distribution of heavy atoms from isotopes in the metabolic networks.
- We perform non-targeted metabolic flux analysis to detect putative metabolite signals that would not be anticipated based on the existing metabolic network models in a specific organism
- Stable isotope labeling is employed to confirm the biosynthetic nature of new metabolites, and be used to outline the active metabolic pathways responsible for the production of new or known metabolites
Network-wide pathway elucidation
Stable isotope labeled metabolite analysis enables the identification of metabolic pathways, providing direct information on novel metabolic pathways. By combining untargeted (or semitargeted) metabolomics with stable isotope labeling, Creative Proteomics has developed a diversity of tools for network-wide pathway elucidation, which in turn accurately identifies and quantifies hundreds of known metabolites. This approach provides the potential for discovering new metabolic pathways and reveals important information about the active pathways of organisms with multiple possible carbon sources.
- Our workflow
The network-wide isotope-labeled metabolomics data can be gained by collection of time-course data, to trace the incorporation of metabolites or by using partially labeled or mixed labeled/unlabeled isotopic precursors
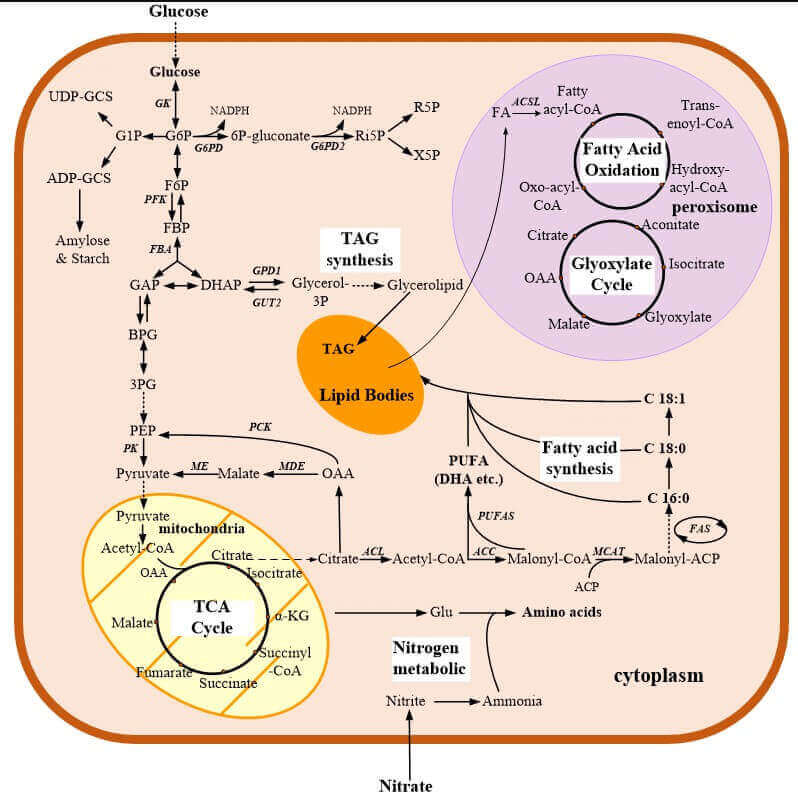 Figure 2. Metabolic pathways in C. cohnii. (Yali, B.; Fangzhong, W. 2019)
Figure 2. Metabolic pathways in C. cohnii. (Yali, B.; Fangzhong, W. 2019)
Features of Our MFA Platform
- Developed based on the most updated knowledge of biology, bioinformatics and software development
- Widely applicable to a wide range of metabolic system
- Professional bioinformatics teams & personalized bioinformatics analysis services.
- Advanced instrument platform
- Integrated quantitative methodologies and comprehensive solutions for metabolomics
Based on high-performance quantitative techniques and advanced equipment, Creative Proteomics has constantly updating our metabolic flux analysis platform and is committed to offering professional, rapid and high-quality services of metabolic pathways study at competitive prices for global customers. Our personalized and comprehensive services can satisfy any innovative scientific study demands, please contact our specialists to discuss your specific needs. We are looking forward to cooperating with you!
References
- Zhiyong, W.; et al. Mechanistic insights into manganese oxidation of a soil-borne Mn(II)-oxidizing Escherichia coli strain by global proteomic and genetic analyses. Scientific Reports. 2017. 7: 1352.
- Yali, B.; Fangzhong, W. Omics Analysis for Dinoflagellates Biology Research. Microorganisms. 2019. 7(9): 288.