Metabolic flux analysis (MFA) has been widely applied to investigate the actual intracellular reaction rates of living microorganisms. MFA utilizes mathematical models to quantitatively infer metabolic fluxes from measurements of multi-step experimental computational protocols. In recent decades, inspired by the diversity of biological problems and driven by the achievements of various experimental analysis, the scope of MFA techniques has been expanded. Isotope-based MFA is the most powerful tool for in vivo metabolic flux quantification with the highest accuracy.
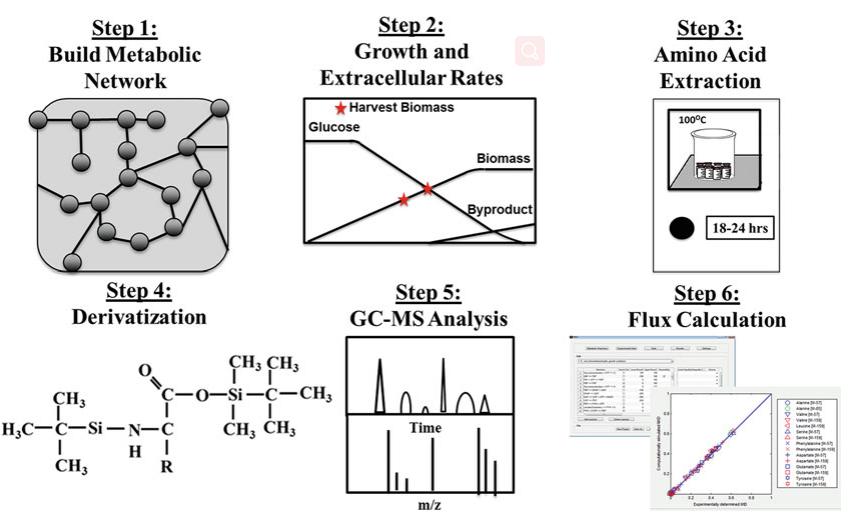 Fig.1 13C-metabolism analysis workflow. (Hollinshead, Whitney, Lian He, and Yinjie J. Tang, 2019)
Fig.1 13C-metabolism analysis workflow. (Hollinshead, Whitney, Lian He, and Yinjie J. Tang, 2019)
Integrating 13C metabolic flux analysis with metabolic engineering
Microorganisms, including a variety of bacteria, have been widely used to produce unique bioactive compounds, playing an important role in the development of the chemistry of natural products, medical therapeutics, and energy industries. With the rapid development of metabolic engineering and synthetic biology, microbial metabolites have been shown to be effective antibacterial agents, enzyme inhibitors, antitumor agents, and biofuels, among others. However, engineered microorganisms are characterized by the high complexity of cell metabolism. Many biosynthetic chemicals are not commercially available on an industrial scale due to low production levels and unsatisfactory yields and productivity. In order to achieve high levels of production of target chemicals, the development of novel strategies in metabolic engineering is essential. 13C-MFA has been widely recognized as one of the most important tools for identifying complex microbial metabolism and developing novel metabolic engineering strategies.
Our services
Based on advanced mathematical algorithms and state-of-the-art instrumentation platforms, we can help our customers to achieve an accurate and quantitative analysis of metabolic fluxes for various bacteria. Our bacterial MFA service is mainly based on parallel 13C tracer studies with GC-MS analysis of amino acids, sugars, and sugar derivatives, optionally complemented by NMR data from amino acids. In addition, we provide other MFA methods for the analysis of the metabolism of bacteria, including 13C isotopic non-stationary metabolic flux analysis (13C-NMFA or INST-MFA) and flux balance analysis (FBA). In general, our services include five major steps: experimental design, cell cultivation, labeling experiment, isotopic analysis of metabolites, and related pathway and flux analysis. We are able to help researchers and professionals to study the following industrial microorganisms, including but not limited to:
Cyanobacteria have received much attention as green catalysts for special biotechnologies because of their metabolic flexibility, single-cell anatomy and high photosynthetic efficiency. They utilize both sugars and carbon dioxide while harnessing the energy of sunlight. This photoheterotrophic mode allows for rapid growth and high cell density, opening up the prospect of sustainable biomanufacturing.
- Escherichia coli (E. coli)
E. coli has been widely used not only as a model microorganism for various biochemical and biotechnological studies, but also as an excellent industrial model microorganism due to its clear genetic background, ease of cultivation, fast anaerobic growth rate, and ability to use a variety of substrates. E. coli is an important strain that cannot be ignored in the field of metabolic engineering.
- Bacillus subtilis
- Corynebacterium glutamicum
As a flexible and powerful method to elucidate intracellular metabolic rewiring, Isotope-based MFA has been widely used in the model and non-model microorganisms and has revealed many metabolic "mysteries" within wild-type, evolved, and engineered strains. In conjunction with multiple MFA methods, we are committed to revealing these biological insights to facilitate the development of rationally designed metabolic engineering strategies that can further enhance biochemical production.
References
- Guo, Weihua, Jiayuan Sheng, and Xueyang Feng. "13C-metabolic flux analysis: an accurate approach to demystify microbial metabolism for biochemical production." Bioengineering 3.1 (2015): 3.
- Hollinshead, Whitney, Lian He, and Yinjie J. Tang. "13 C-Fingerprinting and Metabolic Flux Analysis of Bacterial Metabolisms." Microbial metabolic engineering: methods and protocols (2019): 215-230.