4D-DIA Quantitative Proteomics Analysis
The fusion of 4D separation technology and DIA mode in proteomics research, known as 4D-DIA proteomics, represents a significant expansion in both the scope and depth of proteomics studies. Drawing upon years of dedicated expertise in proteomics, Creative Proteomics is poised to offer top-tier 4D-DIA proteomics services designed to accelerate and enrich your protein research endeavors.
What is 4D-DIA Quantitative Proteomics
4D-DIA quantitative proteomics stands as a cutting-edge method in the realm of MS-based proteomics. This innovative approach expands upon traditional 3D separation, which considers factors such as retention time, m/z, and intensity, by introducing a fourth dimension: ion mobility separation. The integration of ion mobility separation into proteomics not only accelerates scanning speed but also enhances detection sensitivity, ultimately amplifying the performance metrics of proteomics analysis. The DIA data acquisition mode represents a key feature of 4D-DIA quantitative proteomics. It conducts a comprehensive analysis of all ions falling within a designated m/z using MS2. The advantages of DIA are manifold, leading to enhanced coverage, accuracy, and reliability in proteomics analysis. It facilitates a more extensive assessment of protein content within a sample, significantly boosting the comprehensiveness and precision of the results. 4D-DIA quantitative proteomics emerges as a robust methodology that marries the benefits of 4D separation technology with the DIA data acquisition mode. This synergy equips researchers to conduct thorough and in-depth investigations of complex proteomes, providing a holistic and precise portrait of the proteome. Consequently, this approach delivers invaluable insights into cellular processes, mechanisms underlying diseases, and the identification of potential biomarkers.
The Principle of 4D-DIA Proteomics
Ion mobility refers to the velocity at which ions traverse a defined gaseous region under the influence of a unit electric intensity. It allows the separation of peptides co-eluting with similar m/z based on their ion shape and cross-sectional characteristics, resulting in the generation of unique MS2 data. Ion mobility exploits differences in ion flight velocity within an airflow, which is determined by the size and shape of the ions. By introducing an opposing electric field that counteracts the ions' motion, it becomes possible to capture the ions at specific locations where the two opposing forces balance. By modulating the electric field strength, ions can be selectively released in a sequential manner, enabling efficient separation.
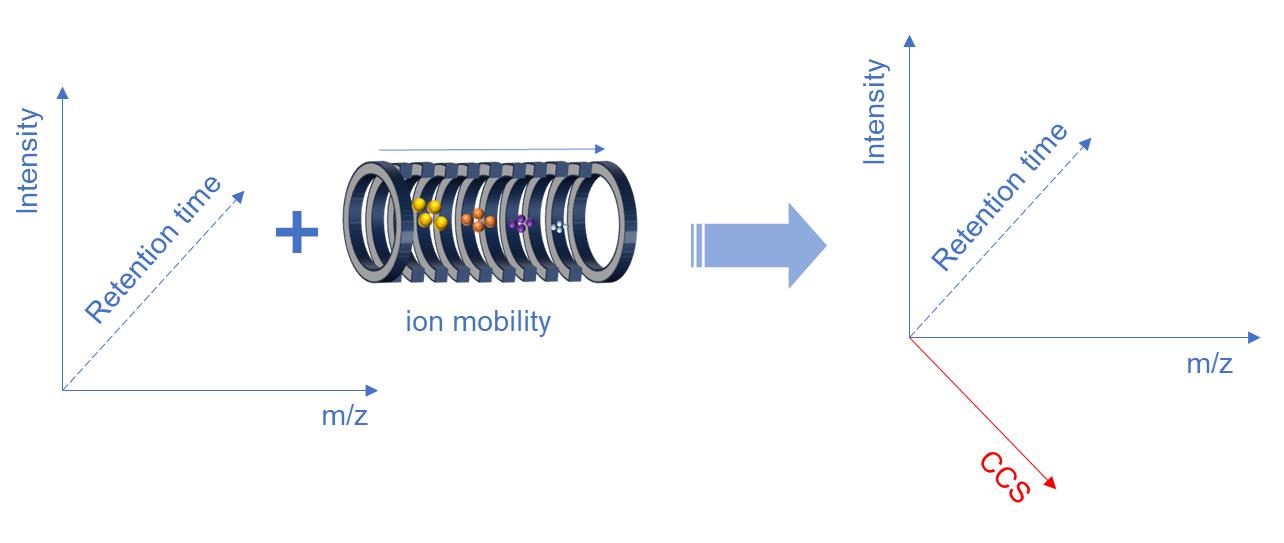
Fig. 1. Schematic of 4D separation technology
Applications of 4D-DIA Proteomics
4D-DIA proteomics offers a variety of valuable applications, and its comprehensive and in-depth analytical capabilities are ideally suited to life science research in medicine, agriculture, the environment, zoology and botany, and more. The applications include pathology research, biomarker discovery, drug target screening, crop improvement, disease-causing mechanisms and drug resistance research, drug response protein expression profiling of knockouts/overexpression, and proteomic library construction of species, etc.
Our 4D-DIA Quantitative Proteomics Analysis Service
Combining the advantages of 4D separation technology and DIA data acquisition modes, 4D-DIA proteomics increases proteomics analysis depth and coverage. It enables improve the speed of scanning and detect more proteins with fewer samples required while ensuring accuracy and reproducibility. Creative Proteomics provides considerate and high quality 4D-DIA quantitative proteomics services. In addition, DIA quantitative proteomics analysis is also available.
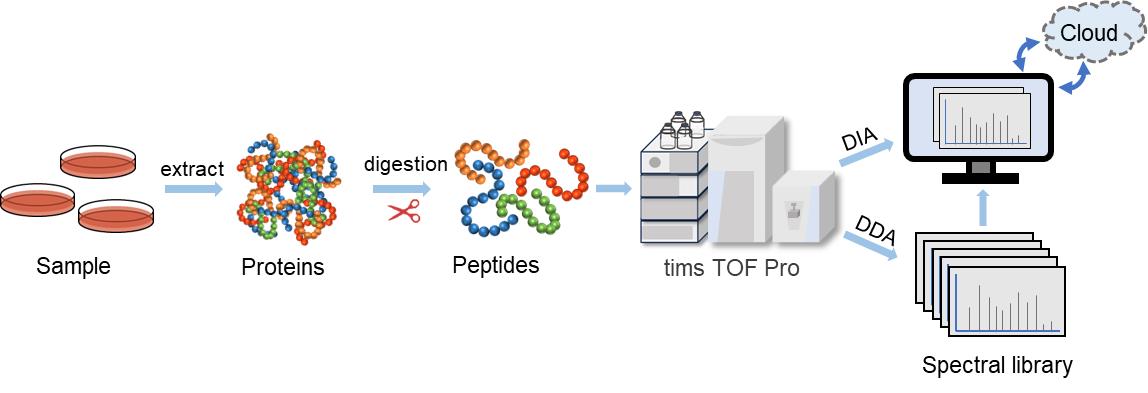
Fig. 2. 4D-DIA Workflow
Advantages of 4D-DIA Proteomics
- A faster scanning speed and a higher detection sensitivity, as well as shorter detection cycles.
- Increased coverage and depth of detection, more comprehensive protein detection.
- Data that is more reliable and accurate.
- Small sample requirements.
Creative Proteomics provides comprehensive and high-quality proteomics analysis services. If you are interested in 4D-DIA quantitative proteomics analysis, please contact us for more details.
Reference
- Chen M.; et al. High-Coverage Four-Dimensional Data-Independent Acquisition Proteomics and Phosphoproteomics Enabled by Deep Learning-Driven Multidimensional Predictions. Analytical Chemistry. 2023, 95(19): 7495-7502

