4D Label-free Quantitative Proteomics Analysis
4D label-free quantitative proteomics stands as cutting-edge, label-free technology built upon the foundation of the timsTOF Pro ion mobility platform. Its innovative approach represents a significant evolution of traditional label-free quantitative proteomics, seamlessly integrating 4D separation technology. Creative Proteomics, drawing upon its extensive dedication to proteomics research spanning numerous years, is uniquely poised to offer expert 4D label-free quantitative proteomics services, effectively bolstering your protein research endeavors.
What Is 4D Label-free Quantitative Proteomics
In protein research, the limitations of traditional proteomics techniques, such as the sensitivity and scanning speed of MS, often fall short of scientific expectations. Consequently, many researchers have turned to labeled quantitative methods. However, with the advent of the timsTOF Pro's ion mobility platform and the ddaPASEF (data-dependent acquisition parallel accumulation-serial fragmentation) mode, 4D label-free proteomics has emerged as a transformative approach. Label-free proteomics compares the relative abundance of proteins in different biological samples without the use of stable isotope labeling or chemical labeling. 4D separation technology identifies low abundance protein signals that are masked under 3D conditions by separating ion mobility. The combination of the two significantly improves coverage, sensitivity, accuracy, and throughput, while also saving expensive isotope costs. It makes cost-effective quantitative proteomics prote.
The Principle of 4D Label-free Quantitative Proteomics
It performs quantitative analysis via the ddaPASEF data acquisition mode. In the ddaPASEF method, multiple precursor ions are stored in an ion mobility tube and then sequentially fragmented, enabling hundreds of MS/MS detections per second. At the same time, it can measure the ion mobility information of peptide segments. 4D label-free quantitative proteomics significantly improves protein detection speed and coverage with higher sensitivity and speed than conventional techniques. It is a new protein analysis method enhanced by the traditional unlabeled method for higher performance.
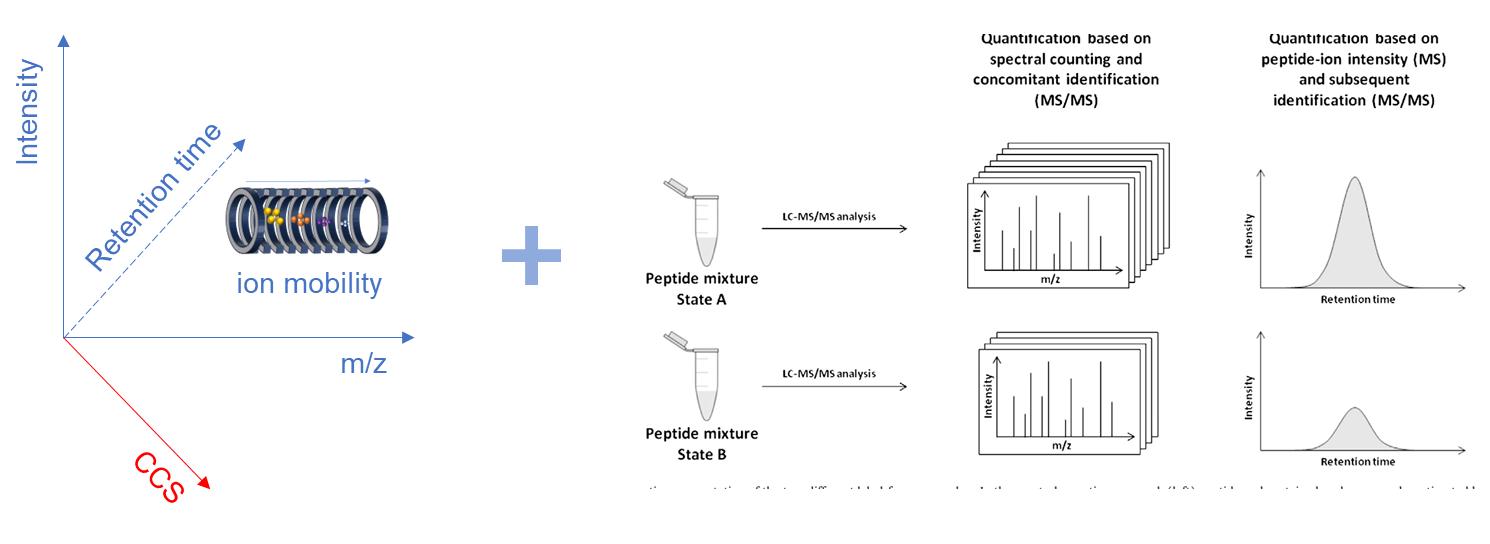
Fig. 1. Schematic of 4D label-free quantitative proteomics (Megger, D. A., et al., 2013)
Applications of 4D Label-free Quantitative Proteomics
The versatility of 4D label-free quantitative proteomics extends across a broad spectrum of scientific domains, including medicine, animal biology, plant biology, and microbiology. This innovative methodology finds application in a multitude of areas, such as pathology, drug mechanism, disease typing, biomarker screening, drug targeting, screening of special functional proteins, plant stress resistance, reproductive development, crop improvement, microbial pathogenicity and drug resistance, etc. In essence, it serves as a versatile tool with applications spanning various scientific disciplines. Its ability to provide comprehensive insights into the proteomics landscape makes it invaluable for advancing research and addressing complex biological questions in medicine, agriculture, and microbiology.
Our Service
Creative Proteomics provides high-quality 4D label-free proteomics services with timsTOF Pro. With high ion utilization and accuracy, it enables higher sensitivity, coverage and throughput through less sample volume and faster scanning speed.
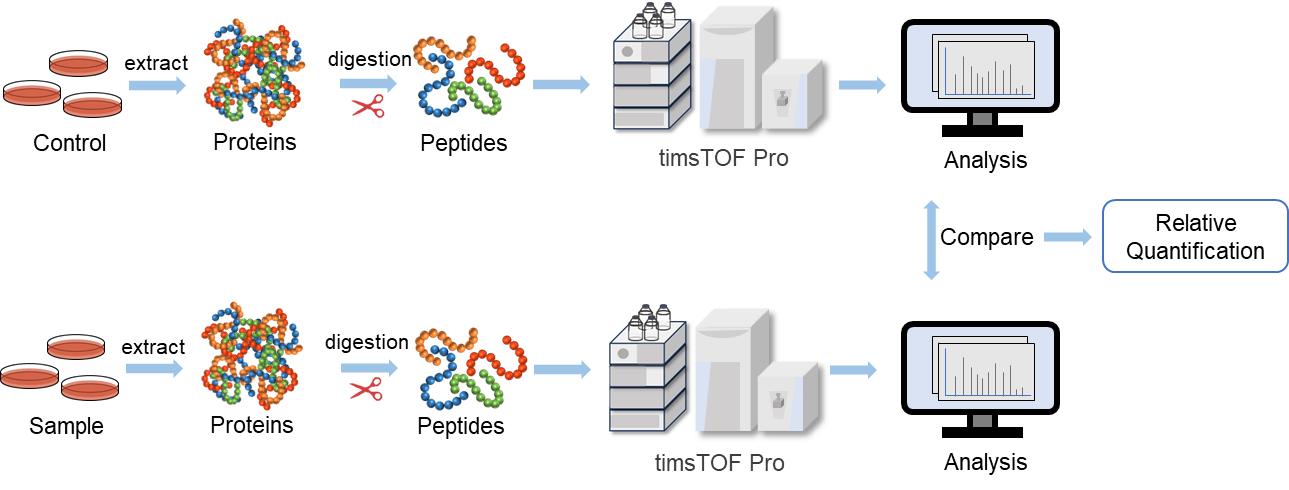
Fig. 2. 4D label-free quantitative proteomics workflow
Advantages of 4D Label-free Quantitative Proteomics Analysis
- Incorporation of ion mobility information, enhancing data quality.
- Exceptional coverage, accuracy, stability, and reliability in protein quantification.
- Reduced sample volume requirements, making it particularly suitable for micro samples.
- High throughput enabled by rapid detection speeds.
Creative Proteomics stands as a trusted source for high-quality analysis services. If 4D label-free quantitative proteomics analysis aligns with your research objectives, please contact us for further details. Together, we can unlock the full potential of this transformative technology, advancing our understanding of proteins and their roles in diverse biological contexts.
References
- Meier, F.; et al. Online Parallel Accumulation-Serial Fragmentation with a Novel Trapped Ion Mobility Mass Spectrometer. Mol Cell Proteomics. 2018, 17(12): 2534-2545
- Megger, D. A.; et al. Label-free quantification in clinical proteomics. Biochim Biophys Acta. 2013, 1834(8): 1581-90

