Protein-RNA Interaction Analysis Service
Protein-RNA interactions play a key role in biological processes, and the precise resolution of them is essential for in-depth studies of protein functions and RNA regulatory mechanisms. As a company that has specialized in proteomics research for many years, Creative Proteomics can provide professional protein-RNA interaction analysis service to facilitate your protein research.
What is Protein-RNA Interaction
Interactions between proteins and RNA are essential in cellular life activities and cellular homeostasis. RNA binding protein (RBP) is a powerful and extensive regulatory factor, and most RNA works when bound to it. RBP is involved in many regulatory processes such as RNA synthesis, splicing, modification, translocation, translation and intracellular localization, and affects mRNA stability, lncRNA activity, miRNA silencing complex formation, etc. Relatively, RNA also regulates RBP's activity, localization, or interaction with other proteins, affecting various cellular processes mediated by it. The protein-RNA interaction is essential for normal cellular life activities.
The Principle of Protein-RNA Interaction Analysis
Currently, there are two main categories of methods for analyzing protein-RNA interactions. One is centered on the target RNA and detects proteins bound to it, such as RNA pull-down and CHIRP, and the other is centered on the target protein and detects RNA bound to it, like RIP. Each method has unique strengths and limitations, therefore choosing the optimal method to solve the biological problem is essential.
RNA pull-down is a technical tool for research into protein-RNA interactions. Its principle is to use an in vitro transcriptionally synthesized RNA probe labeled with biotin, which is incubated in a cytoplasmic protein extraction solution to form an RNA-protein complex. The complex binds to streptavidin-labeled magnetic beads, which separates it from other components in the incubation solution. After elution of the complex, protein-RNA interactions are detected by WB assays or combined with MS to screen for unknown proteins bound to RNA. For mutually verifying the reliability of experimental results, it is often used with RIP.
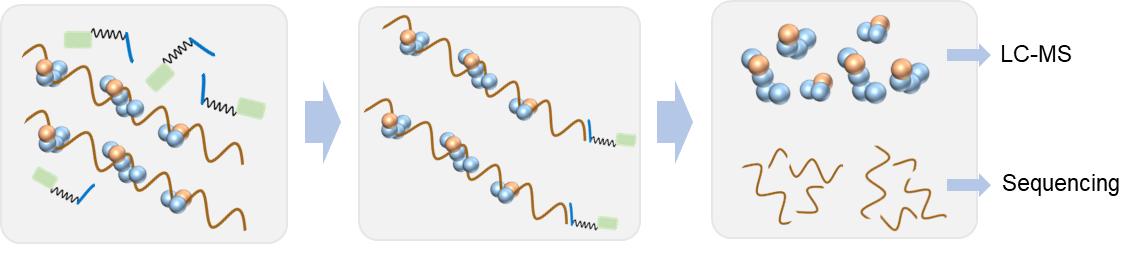
Fig. 1. Schematic of protein-RNA interaction analysis
CHIRP is a method for detecting RNA-bound DNA and protein interactions in vivo. The method is to design a biotin probe that is complementary to the target RNA sequence to pull down the target RNA. And then the DNA chromosome fragment interacting with the target RNA is indirectly attached to the magnetic beads. After elution, measure the target RNA and DNA by qRT-PCR, sequencing, etc., and then determine the proteins by WB experiment or mass spectrometry.
RIP is also a method for researching protein-RNA interactions on similar principles, but it is protein-centered. It uses antibodies against target proteins to precipitate the corresponding protein-RNA complexes, and after isolation and purification, the RNA in the complexes can be determined by qRT-PCR, sequencing, etc. Experimental results are often statistically verified by using it in conjunction with RNA pull-down.
Table 1. Differences in protein-RNA interaction analysis methods.
| Method | RNA pull-down | CHIRP | RIP |
| Classification | RNA-centered | RNA-centered | protein-centered |
| In Vivo/Vitro | In vitro | In vivo | In vitro |
| Probe | Target RNA, length limited | DNA/RNA probes complementary to target RNA, unaffected by length | Antibody |
| Research object | RNA-protein | RNA-protein, RNA-DNA | Protein-RNA |
Our Service
Creative Proteomics provides considerate and high-quality protein-RNA interactions analysis services. And we offer all the analytical methods described above and can provide you with the optimal solution for your research requirements. In addition, we provide protein-protein interaction analysis service, protein-metabolite interaction analysis service and protein-DNA interaction analysis service.
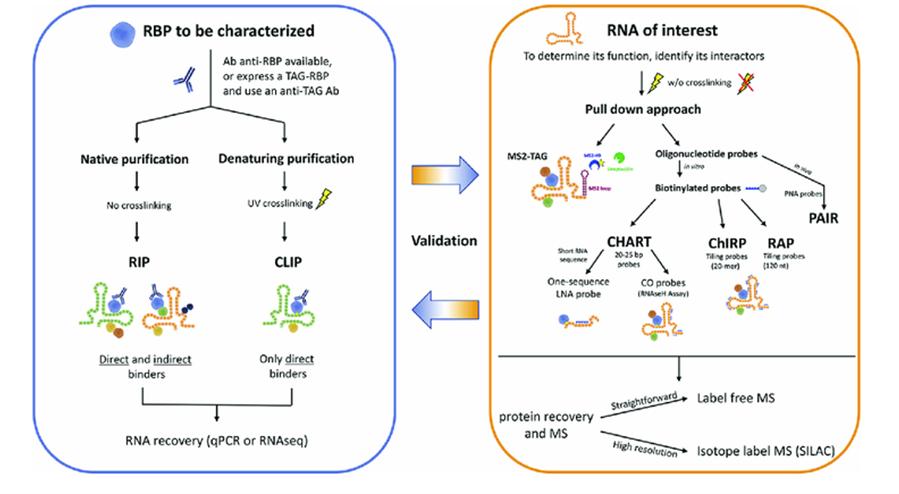
Fig. 2. Workflow of protein-RNA interaction analysis (Barra J, 2017)
Applications of Protein-RNA Interaction Analysis
The biological functions of many proteins and RNAs are also affected by protein-RNA interactions. For example, some enzymes, transcription factors, regulatory proteins, etc., their functions require binding to RNA. Meanwhile, RNA can also be a target for proteins and applied to drug development or the treatment of diseases. Protein-RNA interactions are one of the most fundamental biochemical processes in living systems. The study of them contributes to a better description of biological processes and provides new ideas and methods for biomedical research.
Advantages of Protein-RNA Interaction Analysis Service
- It studies intracellular RNA-protein binding.
- It studies RBP interactions with non-coding RNAs.
- It can map RNA-RBP interactions.
- It can be used to validate experimental results in both directions.
Creative Proteomics provides comprehensive and high-quality proteomics analysis services. If you are interested in protein-RNA interaction analysis, please contact us for more details.
References
- Mattay J. Current Technical Approaches to Study RNA–Protein Interactions in mRNAs and Long Non-Coding RNAs. BioChem. 2022; 3(1):1-14
- Barra J, Leucci E. Probing Long Non-coding RNA-Protein Interactions. Front Mol Biosci, 2017, 4: 45

