Olink Proteomics Analysis Service
Olink Proteomics stands at the forefront of cutting-edge proteomics technologies, offering a unique approach to studying proteins through its Proximity Extension Assay (PEA) technology. With a wealth of expertise, Creative Proteomics extends the top-tier olink proteomics service to provide a valuable perspective on your protein-focused research.
What is Olink Proteomics
Olink Proteomics relies on PEA technology, which is capable of measuring multiple proteins simultaneously with high sensitivity and specificity. Each antibody pair of PEA is attached to a unique oligonucleotide probe, which is complementary in close proximity when the antibody binds to the target protein, forming a nucleotide chain. Using this dual recognition binding principle, PEA converts protein information into nucleic acid signals, greatly enhancing detection specificity and minimizing background noise. It accurately detects low-abundance proteins and can quantify up to 92 protein biomarkers simultaneously, and has become a leading method for multiplexed protein analysis.
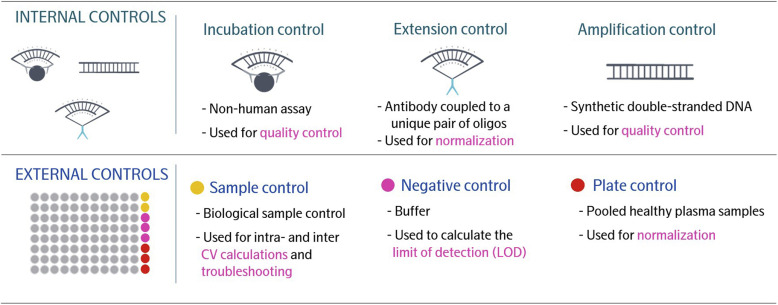
Fig. 1 Schematic of PEA technology (Wik, L., et al.; 2021)
PEA is a targeted proteomics quantification method that combines antibody-based immunoassays with PCR or Next Generation Sequencing (NGS) technology. It begins by designing antibodies against target proteins so that complementary oligonucleotide probes are attached to each antibody. Then, after the antibody specifically binds to the target protein, the probe pairs on the antibody undergo proximity hybridization to form a PCR template, followed by amplification of the sample using primers. Finally, quantification is achieved by qPCR or NGS. Through this process, the protein concentration signal is converted to a nucleic acid signal, enabling the detection of low abundance proteins.
Our Olink Proteomics Service
Olink is dedicated to providing researchers with cutting-edge platforms for ultra-sensitive detection, high-throughput analysis, targeted biomarker screening, and drug target validation. By integrating traditional proteomics methodologies, Olink facilitates the translation of clinical research into practical applications, thereby establishing a robust framework for precision medicine centered on proteomics.
Creative Proteomics offers specialized technical services utilizing Olink's Target and Explore series, leveraging advanced platforms to enable precise quantification of 48 to 3,072 biomarkers. This significantly enhances research efficiency and accelerates the translation of scientific discoveries into tangible outcomes.
Olink Product Overview: Olink Target Series
The Olink Target series encompasses a comprehensive range of disease-specific panels designed to support precision proteomics research. Below is a detailed list of available panels:
Disease-Specific Panels:
- Cardiovascular Disease-II Panel
- Cardiovascular Disease-III Panel
- Oncology-II Panel
- Oncology-III Panel
- Neurology Panel
- Inflammation Panel
- Immune-Oncology Panel
Biological Process (BP)-Related Panels:
- Cardiac Metabolism Panel
- Cellular Regulation Panel
- Developmental Biology Panel
- Immune Response Panel
- Metabolism Panel
- Neuroexploration Panel
- Organ Injury Panel
These panels are tailored to address specific research needs, enabling the identification and quantification of biomarkers associated with various diseases and biological processes. By utilizing Olink's state-of-the-art technology, researchers can achieve unparalleled precision and efficiency in their proteomics studies.
Advantages of Our Services
High-Throughput and Unbiased Detection:
Our platform enables the parallel detection of up to 3,072 protein biomarkers, providing comprehensive coverage of all major biological signaling pathways.
Exceptional Sensitivity:
The detection sensitivity for target proteins reaches the femtogram per milliliter (fg/mL) level, allowing for the simultaneous quantification of hundreds of disease-associated low-abundance proteins at the omics level. This capability is particularly advantageous for analyzing key low-abundance biomarkers in diverse biological fluids, including plasma, serum, cerebrospinal fluid, tumor interstitial fluid, and urine.
Broad Dynamic Detection Range:
The linear detection range spans 10 orders of magnitude, ensuring precise quantification of target proteins across a wide spectrum of abundances, from highly abundant to extremely low-abundance molecules.
Outstanding Specificity:
Leveraging the unique probe design of the Proximity Extension Assay (PEA) technology, our platform effectively mitigates non-specific binding interference commonly encountered in multiplex antibody assays. This ensures that each detected target is a well-defined biomarker, addressing the challenges of non-specificity faced by traditional proteomics methods.
Minimal Sample Requirements:
Each assay requires only 1 microliter of sample, minimizing the volume needed for analysis while maintaining high precision.
Data Reliability:
We have established a rigorous internal quality control system, incorporating multi-dimensional quality control metrics to ensure the reproducibility and accuracy of the detection data. This robust framework guarantees the reliability of our results, supporting high-quality scientific research.
Workflow of Olink Proteomics Analysis Service
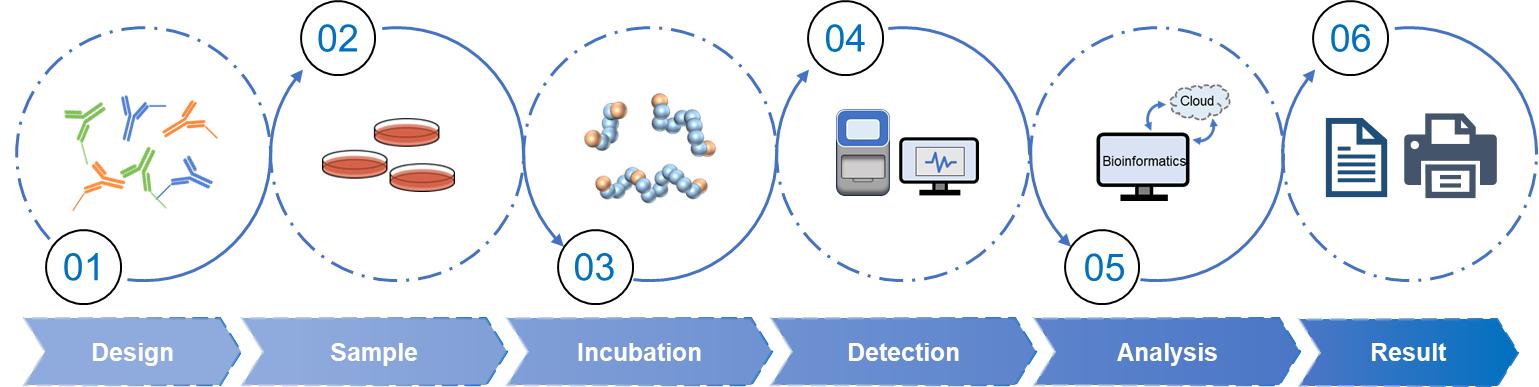
Fig 2. Workflow of olink proteomics
Applications of Olink Proteomics
Medicine
It plays an important role in identifying and validating biomarkers, contributing to the development of medical diagnostics and personalized medicine.
Disease Research
Its ability to measure multiple proteins in a single sample is particularly important for studying complex diseases caused by multiple factors.
Biology Research
As a result of its versatility, it is able to provide insight into the intricate protein networks and signaling pathways that operate in various biological systems.
Sample Requirements
Compatible Sample Types:
Our platform is compatible with a wide range of biological samples, including:
- Standard Samples: Serum and plasma.
- Non-Standard Samples: Cerebrospinal fluid, aqueous humor, urine, tissue lysates, cell culture supernatants, cell lysates, microvesicles/exosomes, interstitial fluid, microdialysates, fine-needle biopsy tissues, dried blood spots, synovial fluid, saliva, and others.
Standard Sample Preparation:
For serum and plasma, no pre-experimental validation is required. Samples can be directly prepared and processed for formal analysis according to the provided guidelines. (Refer to the Blood Serum Sample Handling Protocol for detailed instructions.)
Non-Standard Sample Preparation:
Non-standard samples (excluding serum and plasma) require pre-experimental validation. For each sample group, please provide 3–4 preliminary samples with the following specifications:
- Volume: 50–60 µL per sample.
- Protein Concentration: 0.5–1 µg/µL.
Frequently Asked Questions (FAQ) of Olink Proteomics
-
Q1: What is Proximity Extension Assay (PEA)?
A1: Olink's Proximity Extension Assay (PEA) technology employs dual antibody recognition to convert protein concentration data into quantifiable DNA molecule counts. This approach uniquely combines specificity and scalability, enabling high-throughput, multiplexed protein biomarker analysis. PEA requires minimal sample volumes and is compatible with nearly all biological sample types without compromising data quality or assay robustness.
-
Q2: Does Olink offer a panel targeting human homologs of proteins in the Target 48 Mouse Cytokine panel to facilitate translational research?
A2: Olink provides the Target 48 Cytokine panel for human proteins, which exhibits 42% overlap with mouse homologs in the Target 48 Mouse Cytokine panel. Additionally, the Olink Flex library covers 75% of the corresponding mouse proteins quantifiable by the Target 48 Mouse Cytokine panel.
-
Q3: Are there panels available for other species, such as mice?
A3: Olink offers two products specifically designed for mouse proteins: the Target 96 Mouse Exploratory and Target 48 Mouse Cytokine panels. The Target 96 Mouse Exploratory panel is also suitable for analyzing rat proteins. While all other Olink assays are developed using antibodies targeting human proteins, some cross-reactivity with homologs from other species may occur. Consequently, a subset of assays in these panels may produce signals in non-human samples, and some customers have successfully used them in animal studies. However, Olink human panels are intended for human samples, and their use in non-human species is at the user's discretion.
-
Q4: Are there biomarkers common to both the Target 48 Mouse Cytokine and Target 96 Mouse Exploratory panels?
A4: Yes, the Target 96 Mouse Exploratory and Target 48 Mouse Cytokine panels share 14 common biomarkers. Results from both panels demonstrate strong correlation when analyzing the same sample.
-
Q5: Why are some proteins in healthy controls poorly detectable?
A5: Certain proteins exhibit a wide range of expression levels in vivo, making it challenging to develop multiplex assays that simultaneously measure proteins in both healthy and diseased individuals. For example, the CVD panel is optimized to detect elevated protein levels associated with cardiovascular disease. If low detectability is observed, non-parametric statistical methods (e.g., detected vs. undetected in groups) can be employed for analysis.
-
Q6: Can I obtain FASTQ data and perform my own analysis?
A6: Traditional DNA sequencing and NGS sequencing for Olink Explore differ fundamentally. Traditional sequencing, used for applications like whole-genome sequencing and mutation screening, involves identifying unknown DNA sequences mapped to a reference genome. In contrast, Olink Explore sequencing counts predefined DNA sequences, with counts directly correlating to protein concentrations. The workflow generates BCL files, which are converted to COUNTS files, and finally to NPX files containing protein concentration data. Olink provides only NPX data, as BCL files contain proprietary molecular design information.
-
Q7: What is the Sample Control in Olink Explore, and how is it used in data analysis?
A7: The Sample Control consists of pooled plasma samples. These serve as external controls to assess intra- and inter-plate precision. Sample Controls are run in triplicate on Explore HT and in duplicate on Explore 384/3072. For Explore HT, Sample Controls are included with the kit.
-
Q8: Can Olink provide raw data from NovaSeq output files?
A8: As described in Question 6, Olink Explore sequencing counts predefined DNA sequences, with counts directly correlating to protein concentrations. The workflow generates BCL files, which are converted to COUNTS files, and finally to NPX files. Olink provides only NPX data, as BCL files contain proprietary molecular design information.
-
Q9: What is the Plate Control in Olink Explore, and how is it used in data analysis?
A9: The Plate Control consists of pooled plasma from healthy donors. It is used for data normalization to account for potential variations between runs and plates. Plate Controls are run in quintuplicate on Explore HT and in triplicate on Explore 384/3072.
-
Q10: What is the Negative Control in Olink Explore, and how is it used in data analysis?
A10: The Negative Control consists of buffer and is processed as a normal sample. It monitors background noise from DNA tags not bound to their target proteins. In Explore 384/3072, Negative Controls establish baseline levels and calculate the limit of detection (LOD). In Explore HT, they assess potential contamination. Negative Controls are run in duplicate on Explore HT and in triplicate on Explore 384/3072.
-
Q11: What does LOD mean?
A11: The Limit of Detection (LOD) is the lowest measurable level of a protein, defined as three times the standard deviation of the background signal.
-
Q12: What does NaN mean?
A12: NaN stands for "Not a Number" and is used to replace data values below the LOD.
-
Q13: What is dynamic range?
A13: Dynamic range refers to the region between the Lower Limit of Quantification (LLOQ) and the Upper Limit of Quantification (ULOQ), within which quantification is accurate and precise.
-
Q14: What do LLOQ and ULOQ mean?
A14: The analytical measurement range is defined by the Lower Limit of Quantification (LLOQ) and the Upper Limit of Quantification (ULOQ).
-
Q15: How are biomarkers selected for Olink panels?
A15: When constructing new panels, information is gathered from public bioinformatics databases such as UniProt, Human Protein Atlas, Gene Ontology, and DisGeNET. A combination of known and exploratory biomarkers is selected and evaluated, and antibodies must pass rigorous validation tests. Panel composition is guided by domain experts.
-
Q16: What instruments are required to run Olink Focus, Flex, Target 96, or Target 48?
A16: The Olink Signature Q100 is required for detection and quantification. A validated PCR instrument with a 100 µL reaction volume and a heated lid is also needed for the extension step. Alternatively, the BioMark HD Reader and IFC Controller HX/MX/AX/Juno by Standard BioTools can replace the Olink Signature Q100.
-
Q17: Should I dilute my samples before sending them to Olink?
A17: Generally, no. For dilution panels, Olink dilutes samples as part of the protocol. However, for matrices other than serum or plasma, consult the Olink support team regarding dilution, protein concentration, or cell density (e.g., lysates or cell culture supernatants).
-
Q18: What is NPX Signature?
A18: The Olink NPX Signature is a user-friendly tool for quality control, normalization, and statistical analysis. It validates data quality, normalizes Olink data, and performs statistical analyses.
-
Q19: Are customized panels available?
A19: Olink offers two types of customized panels:
- Olink Flex: Highly flexible, allowing selection of up to 21 proteins per panel. Results are reported in absolute (pg/mL) and relative (NPX) quantification.
- Olink Focus: Customizable through a collaboration with R&D experts, enabling detection of up to 21 user-selected proteins with optional absolute or relative quantification.
-
Q20: Is there software for preprocessing and analyzing data from Olink Target, Flex, and Focus?
A20: Olink Proteomics has developed the NPX Signature software for data preprocessing and analysis. This tool facilitates data import, quality control, normalization, and statistical analysis, providing standardized protein expression (NPX) or pg/mL values.
-
Case Study
Data-Driven Discovery and Validation of Circulating Blood-Based Biomarkers Associated with Prevalent Atrial Fibrillation
Background:
Atrial fibrillation (AF) is one of the most common causes of arrhythmia or irregular heart rhythm. Patients with AF are at a significantly higher risk of developing blood clots and experiencing strokes. However, this serious condition is often detected too late. Early screening of high-risk individuals is critical to enable timely initiation of anticoagulant therapy for prevention.
Currently, electrocardiography (ECG) is the only method for detecting AF. Clinical risk factors associated with AF, such as advanced age, prior stroke history, obesity, hypertension, diabetes, and heart failure, are used to identify subpopulations suitable for ECG screening. However, these risk factors, either individually or in combination, have limited predictive power, and their assessment requires specialized expertise, posing challenges for effective screening.
Study Design:
To address these challenges, researchers from the University of Birmingham utilized the Olink Cardiovascular panel to analyze blood samples from 638 hospitalized patients collected between 2014 and 2016. The study evaluated seven clinical risk factors alongside cardiovascular biomarkers.
Key Findings:
Biomarker Identification:
Among all biomarkers analyzed, two were most strongly associated with AF:
Brain Natriuretic Peptide (BNP)
Fibroblast Growth Factor-23 (FGF-23)
Predictive Model:
A simple assessment combining three clinical risk factors (age, sex, and body mass index [BMI]) with the two biomarkers (BNP and FGF-23) demonstrated the potential to identify AF patients effectively.
Stratification Potential:
BNP and FGF-23 may also aid in stratifying AF patients based on disease severity or risk.
Publication:
The findings were published in the high-impact journal European Heart Journal (Impact Factor: 22.7) under the title "Data-driven discovery and validation of circulating blood-based biomarkers associated with prevalent atrial fibrillation."
Implications:
This study highlights the utility of blood-based biomarkers, particularly BNP and FGF-23, in improving the identification and stratification of AF patients. The integration of biomarkers with clinical risk factors offers a promising approach to enhance early detection and management of AF, ultimately reducing the risk of stroke and other complications.
References
- Verwer MC, et al. Comparison of cardiovascular biomarker expression in extracellular vesicles, plasma and carotid plaque for the prediction of MACE in CEA patients. Scientific Reports (2023).
- Hyland M, et al. Pro-Inflammatory Priming of Umbilical Cord Mesenchymal Stromal Cells Alters the Protein Cargo of Their Extracellular Vesicles. Cells (2020).
- Cano A,et al. Plasma extracellular vesicles reveal early molecular differences in amyloid positive patients with early-onset mild cognitive impairment. Nanobiotechnology (2023).
- Krishnamachary B, et al. Extracellular vesicle-mediated endothelial apoptosis and EV-associated proteins correlate with COVID-19 disease severity. Journal of Extracellular Vesicles (2021).
- Viktorsson K, et al. Profiling of extracellular vesicles of metastatic urothelial cancer patients to discover protein signatures related to treatment outcome. Molecular Oncology (2022).

