Co-Immunoprecipitation (Co-IP) Service
What is Co-Immunoprecipitation (Co-IP)?
The Co-Immunoprecipitation (Co-IP) technique is widely regarded as one of the most effective methods for studying protein-protein interactions (PPIs) in living organisms under non-denaturing conditions. This approach enables the identification of interactions involving a wide array of protein partners, including complex partners, structural proteins, cofactors, and signaling molecules. Co-IP serves as a critical enrichment strategy for isolating low-abundance target proteins within a complex biological milieu. Following Co-IP, various techniques such as Western Blot (WB) and LC-MS/MS can be employed to identify interacting partner proteins and explore novel functions associated with the bait protein.
Creative Proteomics boasts extensive expertise in proteomics and offers comprehensive Co-Immunoprecipitation (Co-IP) testing services for pharmaceutical companies and research institutions. This advanced technology not only enables the verification of interactions between two known proteins, but also facilitates the identification of unknown proteins that interact with known proteins within a complex biological context.
What is the Difference Between Immunoprecipitation (IP) and Co-IP?
| Aspect | IP | Co-IP |
| Objective | Isolate a single protein of interest | Identify and isolate interacting partners of a protein |
| Interaction Study | Does not study interactions directly | Focuses on protein-protein interactions |
| Target Proteins | Typically one protein | Proteins that interact with the target protein |
| Complexity | Simple, often used for purification or quantification | Complex, requires validation to confirm specific interactions |
| Applications | Protein purification, quantitative analysis | Investigating protein-protein interactions, signaling pathways |
What is the Difference Between Chromatin Immunoprecipitation (ChIP) and Co-IP?
| Aspect | ChIP | Co-IP |
| Purpose | Used to study protein-DNA interactions | Used to study protein-protein interactions |
| Sample Type | Chromatin (DNA-protein complexes) isolated from cells or tissues | Protein lysates or extracts from cells or tissues |
| Target Molecules | Focuses on identifying proteins bound to specific DNA regions | Focuses on identifying proteins bound to other proteins |
| Binding Partners | Captures DNA fragments associated with a specific protein of interest | Captures protein complexes associated with a specific protein |
| Technique | Requires cross-linking (e.g., formaldehyde) to stabilize protein-DNA interactions | Typically does not require cross-linking |
| Output | DNA sequences bound by the target protein, analyzed using qPCR, sequencing, or microarrays | Associated proteins, identified using techniques like Western blotting or mass spectrometry |
| Applications | Epigenetics, transcriptional regulation, and chromatin structure studies | Signaling pathway analysis and identification of protein interaction networks |
Creative Proteomics' Co-IP Service Workflow
Sample Collection and Preparation
Endogenous Interactions : Protein samples are obtained by homogenizing tissues or lysing cells expressing the bait protein to release proteins into solution.
Non-Endogenous Interactions : Purified bait proteins, often tagged for detection, are incubated with cell lysates to study interactions under native conditions.
Antibody Selection
The success of Co-IP heavily relies on selecting an appropriate antibody that will specifically bind to the bait protein. There are two common strategies:
Specific Antibodies : If a specific antibody for the bait protein exists, it can be used to capture the protein along with its interaction partners from the sample.
Tagged Antibodies : If no suitable antibody is available, tagged antibodies (e.g., Myc, HA, or Flag) bind to tags in the bait protein, enabling isolation and capture of interaction partners.
Protein Complex Immunoprecipitation
Incubation : The antibody-bound beads are incubated with the sample, allowing the bait protein to bind with its interaction partners.
Protein Complex Formation : The bait protein, along with the interacting proteins, forms a complex, which is now captured on the antibody-conjugated beads.
Wash and Elution
Washing with buffer solutions removes non-specifically bound proteins and contaminants, reducing background noise and false positives. Multiple wash steps are typically used.
Washing : The antibody-bead-protein complexes are washed under stringent conditions to minimize non-specific binding.
Elution : After washing, the target protein complex is eluted from the beads, typically by changing the buffer conditions (e.g., using low pH, high salt concentration, or denaturing conditions). This step releases the purified protein complex for downstream analysis.
Mass Spectrometry or Western Blot Analysis
Two primary techniques are used to identify and quantify the interacting proteins:
Mass Spectrometry (Co-IP/MS) : Mass spectrometry offers high resolution and sensitivity for identifying and quantifying proteins within the immunoprecipitated complex.
Western Blot (WB) : WB detects specific proteins in the eluted complex by separating them via SDS-PAGE, transferring to a membrane, and probing with antibodies to confirm interactions.
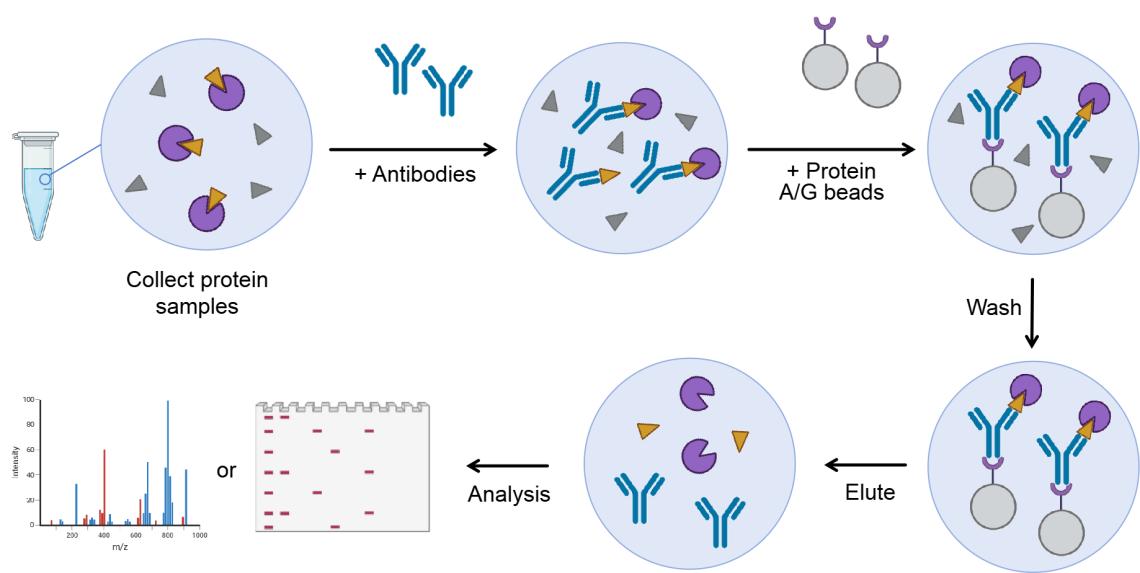
Figure 1. Principle of Co-IP technology.
Why Choose Creative Proteomics for Co-IP Services?
Advanced Technology : We leverage state-of-the-art technologies, including high-resolution mass spectrometry, to deliver cutting-edge data that enhances the quality and accuracy of your research.
Customizable Protocols : Recognizing that every experiment is unique, our Co-IP protocols are fully customizable to align with your specific research objectives and protein targets.
Comprehensive Reporting : Our in-depth reports provide thorough data analysis, allowing you to easily interpret results and make well-informed decisions based on reliable findings.
Global Reach : We extend our services to research institutions, pharmaceutical companies, and biotech organizations around the world, ensuring that our expertise is accessible wherever you are located.
Mass Spectrometry Platform for Co-IP Analysis

Applications of Co-IP Analysis
Protein Complex Identification : Uncover novel protein complexes and their roles in cellular processes.
Signal Transduction Pathways : Investigate signaling events by identifying proteins involved in key cellular pathways.
Drug Target Discovery : Identify potential drug targets by understanding protein interactions in disease models.
Post-translational Modifications (PTMs) : Explore the effects of PTMs on protein function and interactions.
Cancer Research : Examine the role of protein interactions in cancer progression and metastasis.
Simple Requirement for Co-IP Analysis
| Sample Type | Recommended Amount | Note |
| Cell Lysate | 1-5 mg protein | Avoid using harsh detergents; keep samples on ice |
| Tissue Lysate | 50-100 mg tissue | Homogenize tissues in a cold lysis buffer |
| Purified Protein | 0.5-1 mg | Ensure protein purity and concentration |
| Antibody Concentration | 1-5 µg for IP | Optimized for specific protein-antibody interaction |
| Buffer Requirements | RIPA, NP-40, or custom buffer | Use buffer systems tailored to your protein and complex |
FAQ
-
Q1 : What types of reporter systems are available, and how are they read or detected?
A1 : Creative Proteomics offers reporter gene systems like GFP and lacZ, tailored to experimental needs. These reporters are measured using fluorescence or colorimetric assays, depending on the experiment's requirements. GFP enables direct visualization under a fluorescence microscope, facilitating real-time monitoring, whereas lacZ offers a simple colorimetric assay via X-gal staining. These reporter systems deliver precise and quantifiable insights into the strength and specificity of interactions.
-
Q2 : Can Y2H be used for high-throughput screening of large protein libraries?
A2 : Yes, Y2H is especially useful for high-throughput screening. Our platform is designed to handle large-scale interactions, enabling you to screen complex protein libraries efficiently. We can provide comprehensive cDNA libraries or construct custom libraries to fit specific research goals. Our advanced screening platform allows rapid processing of thousands of potential interactions, ensuring that you get comprehensive data in a timely manner.
-
Q3 : How do you handle protein libraries and cDNA construction?
A3 : Creative Proteomics uses a high-quality, well-validated process for constructing protein libraries. We begin with RNA extraction from your target tissues or cells, followed by cDNA synthesis, and then create a plasmid library that includes a wide diversity of potential protein interactions. Our optimized protocols ensure that the library is comprehensive and functional, enabling you to identify a broad spectrum of interactions. Custom libraries can also be designed if you need a specific focus or if you want to use specialized protein sources.
-
Q4 : What validation methods do you offer to confirm the interactions found through Y2H?
A4 : After initial Y2H screening, we offer several validation methods to confirm the interactions detected. These include:
-
Q5 : Can you handle protein interactions in difficult or unconventional conditions (e.g., membrane proteins, post-translational modifications)?
A5 : Yes, Y2H is commonly used for soluble proteins but can also study membrane-bound proteins and proteins with post-translational modifications (PTMs) with some optimizations. For membrane proteins, we use protocols to enhance expression and stability in yeast. Custom modifications can be added to account for PTMs critical to the interactions being studied, ensuring biologically relevant analysis.
Reverse Bait-Prey Testing : Swapping the bait and prey proteins to confirm interaction specificity.
Co-Immunoprecipitation (Co-IP) : This orthogonal technique confirms interactions in a different biological context (in mammalian or yeast cells).
Pull-Down Assays : Another confirmation method that works well for validating protein-protein interactions.
These techniques ensure that the interactions detected by Y2H are biologically relevant and not spurious.
Demo
Demo 1: Effective Identification of Akt Interacting Proteins by Two-Step Chemical Crosslinking, Co-Immunoprecipitation and Mass Spectrometry
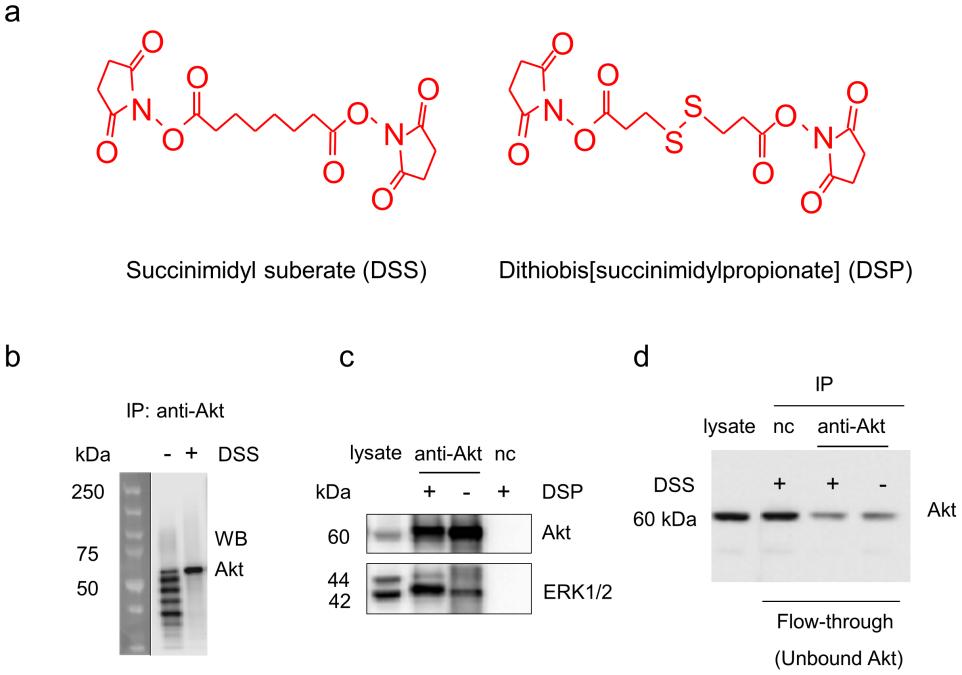
Figure 2. Western blot analysis of Akt and ERK1/2 monitored during co-IP procedures. a) Chemical structures of the crosslinkers used in the study. b) Antibody contamination was eliminated by DSS-crosslinking of Akt antibody to protein A/G agarose beads. c) ERK1/2 co-immunoprecipitation with Akt was significantly improved by DSP-crosslinking. d) DSS-crosslinking did not alter antibody-antigen binding. (Bill X. Huang, et al., 2013)
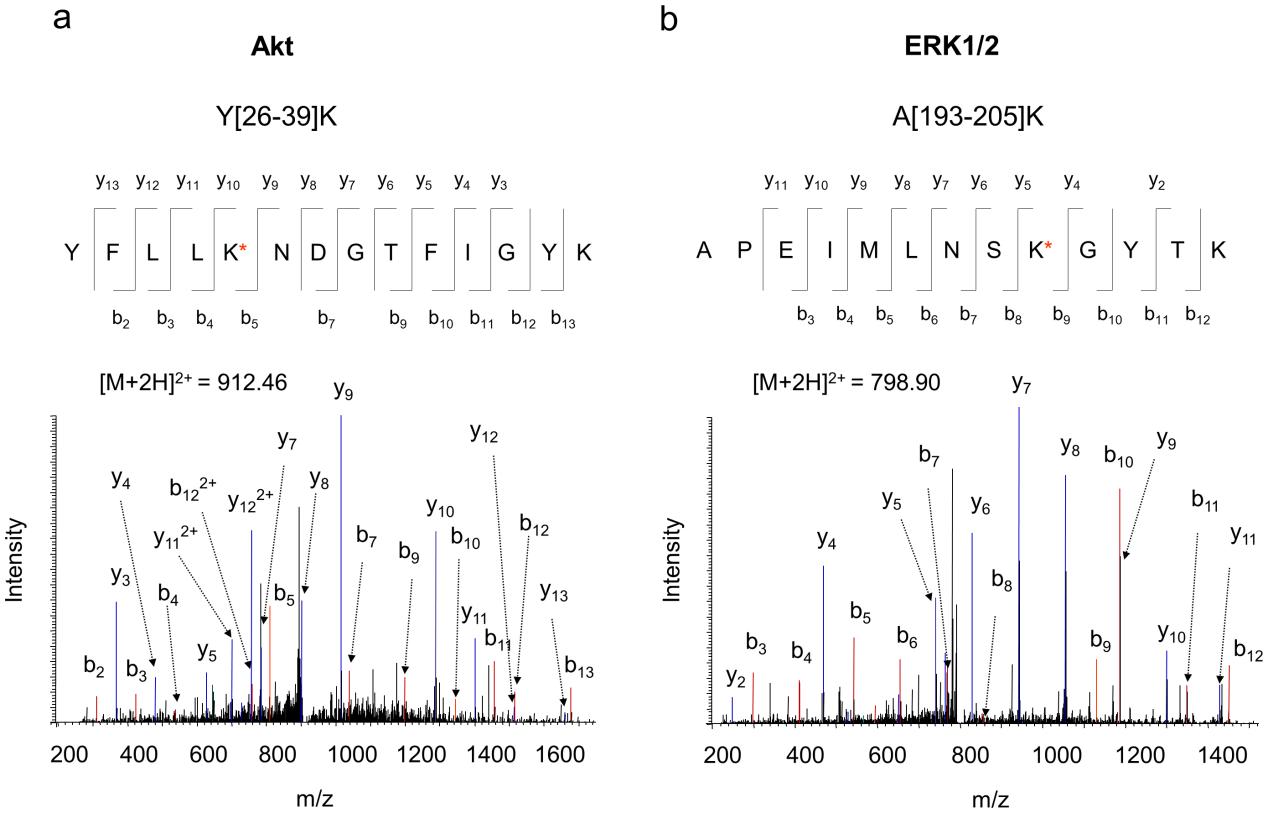
Figure 3. Representative MS/MS spectra of Akt and ERK1/2 peptides obtained from the co-IP complex. MS/MS analysis of the doubly charged ion at m/z 912.46 or 798.90 originated from the peptide segment Y[26]–[39]K of Akt (a), or A[193–205]K of ERK1/2 (b), respectively. The sequence of the peptide was assigned with a single letter abbreviation based on the fragment ions observed for the peptide segments.
-
Case Study
Case: Using yeast two-hybrid system and molecular dynamics simulation to detect venom protein-protein interactions
Abstract
The study shows that MTA1 interacts with the Ku70/Ku80 complex in HCT116 colon cancer cells. This interaction is confirmed both in vitro through Co-IP and in vivo using an in situ detection method. MTA1 and the Ku complex primarily interact in the nucleus, with a weaker presence in the cytoplasm. Furthermore, MTA1 and Ku proteins exhibit synchronized colocalization during the cell cycle, particularly in interphase, prophase, and anaphase, with distinct separations occurring during telophase. The study also shows the MTA1–Ku complex interaction is relevant in the DNA damage response, particularly after UV-induced DNA damage.
Methods
Co-immunoprecipitation (Co-IP) : This technique was used to isolate the Ku70/Ku80 complex using specific MTA1 antibodies.
Fluorescent Microscopy & Co-localization : Fluorescence-based methods were employed to visualize MTA1 and Ku protein interactions during various phases of the cell cycle.
An in situ detection method was used to confirm the in vivo interaction between MTA1 and the Ku complex at different locations within the cell.
Results
Interactome Profiling : Co-IP analysis revealed that the Ku complex (Ku70 and Ku80) was identified as a novel interactor of MTA1 in HCT116 cells, alongside core NuRD components.
In Vitro Co-IP : Co-IP confirmed that MTA1 binds to both Ku70 and Ku80. This interaction was predominantly observed in the nucleus but also occurred in the cytoplasm at a lower level.
DNA-independent Interaction : DNase I treatment did not affect the MTA1-Ku interaction, suggesting that the binding is independent of DNA.
In Situ PLA : PLA analysis confirmed the in vivo interaction between MTA1 and the Ku complex, with interaction sites predominantly in the nucleus, and some in the cytoplasm.
Cell Cycle Dynamics : Fluorescent co-localization showed that MTA1 and Ku proteins colocalized in the nucleus during interphase and moved synchronously through mitosis. At telophase, they were separated and moved asynchronously. The interaction was detected at both interphase and mitosis, particularly at the periphery of chromosomes during mitosis.
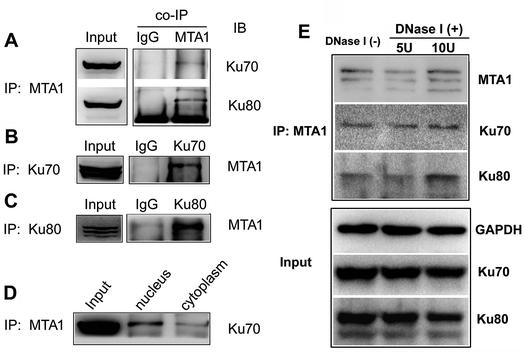
Figure 4. Co-IP analyses on the interaction between MTA1 and Ku70/Ku80. (A) Co-IP analysis performed using MTA1 antibody to pull down Ku70 and Ku80 in whole lysis from HCT116 cells. (B) Co-IP analysis performed using Ku70 antibody to pull down MTA1 in total HCT116 cell lysis. (C) Co-IP analysis performed using Ku80 antibody to pull down MTA1 in total HCT116 cell lysis. (D) The HCT116 nuclear and cytoplasmic cell lysates were used respectively for co-IP analysis. (E) Influence of DNase I treatment on MTA1–Ku interaction.
Related Services
References
- Lin J S, Lai E M. Protein–protein interactions: co-immunoprecipitation. Bacterial protein secretion systems: Methods and protocols , 2017: 211-219. DOI: 10.1007/978-1-4939-7033-9_17
- Liu J, et al. NuRD subunit MTA1 interacts with the DNA non-homologous end joining Ku complex in cancer cells. RSC advances , 2018, 8(61): 35218-35225. DOI: 10.1039/C8RA06907G
- Huang B X, Kim H Y. Effective identification of Akt interacting proteins by two-step chemical crosslinking, co-immunoprecipitation and mass spectrometr. PLoS One , 2013, 8(4): e61430. DOI: 10.1371/journal.pone.0061430

