Yeast Two-Hybrid (Y2H) Service
What is Yeast Two-Hybrid (Y2H)?
The Yeast Two-Hybrid (Y2H) assay is an exceptionally powerful technique for studying protein-protein interactions (PPIs) interactions. Notably, it excels in identifying novel interacting partners by screening a single protein or domain against a library of other proteins. This ability to discover unknown interacting proteins without prior knowledge of their identity is one of the key strengths of the Y2H assay.
As a robust genetic system, Y2H is one of the most standardized methods for mapping PPIs, both at a small scale and in high-throughput formats. During Y2H screening, PPIs are detected through the activation of reporter genes, which respond to a reconstituted transcription factor (TF). This technique is commonly used to test "prey" proteins for interactions with a single "bait" protein or a pool of target proteins. One of the primary advantages of this method is its ability to directly identify interacting protein pairs without the need for additional downstream experiments, as the prey protein is already known and does not require further validation.
Y2H system is widely used to identify novel protein interactions, validate known PPIs, and map interaction networks. It supports research on signaling pathways, biomarkers, and protein complexes. Creative Proteomics provides reliable, cost-effective Y2H services for proteomics, genomics, and drug discovery.
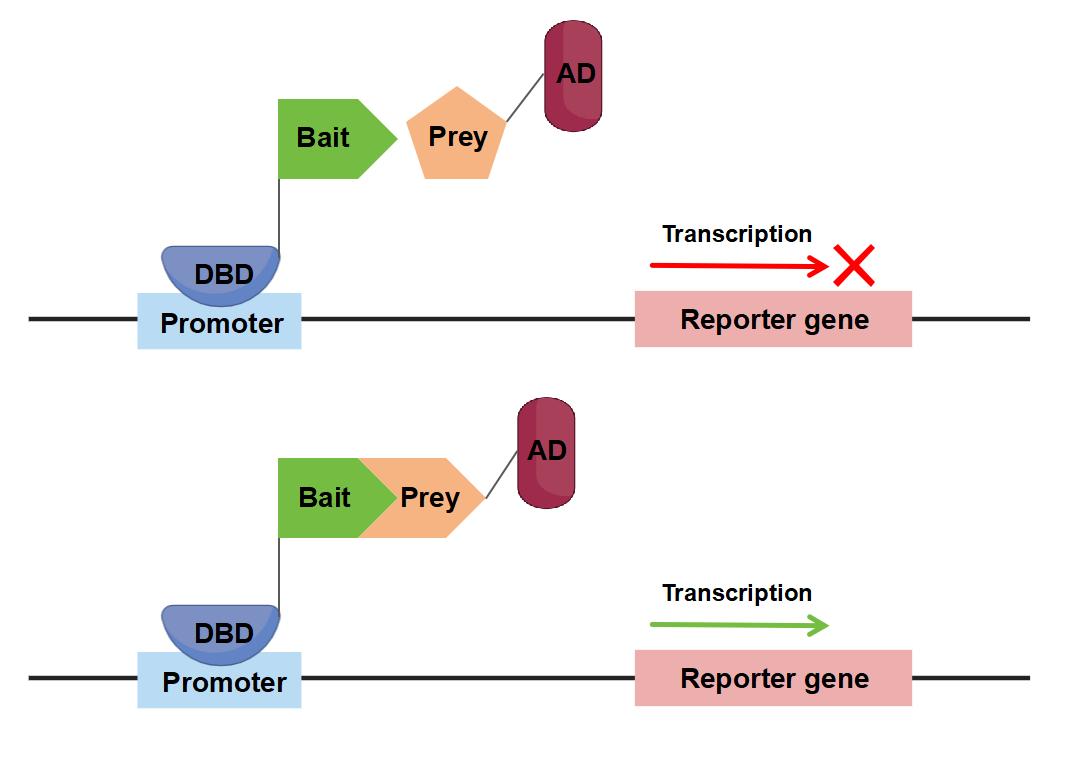
Figure 1. The classical Y2H system for biosensing protein interactions.
What is the Difference Between Y2H and Other Hybrid System?
| Yeast Two-Hybrid (Y2H) | Yeast One-Hybrid (Y1H) | Mammalian Two-Hybrid (M2H) | Bacterial Two-Hybrid (B2H) | Reverse Yeast Two-Hybrid (rY2H) | |
| System Type | Yeast | Yeast | Mammalian Cells | Bacteria | Yeast |
| Interaction Type | PPIs | Protein-DNA Interactions | PPIs | PPIs | PPIs |
| Reporter System | Reconstitution of Transcription Factor (TF) + Reporter Gene | Reconstitution of Transcription Factor (TF) + Reporter Gene | Reconstitution of Transcription Factor (TF) + Reporter Gene | Reconstitution of Transcription Factor (TF) + Reporter Gene | Reporter Gene Expression upon Inhibition of Interaction |
| Sensitivity | High sensitivity to weak or transient interactions | High sensitivity, but limited to DNA-protein interactions | Can study mammalian-specific protein interactions | Suitable for high-throughput screening in bacteria | Useful for studying protein interactions in more complex cellular environments |
| Advantages | High-throughput, comprehensive screening, in vivo context | DNA-protein interaction specificity, in vivo validation | Mammalian environment for protein interaction validation | Fast, cost-effective, easy to handle in bacterial systems | Useful for studying context-specific interactions |
| Applications | Large-scale PPI screening, signaling pathways, gene function discovery | Study of DNA-binding proteins, transcription factors | Study of mammalian-specific PPIs, therapeutic target discovery | PPI discovery in bacterial systems, protein function analysis | Study of interactions in living cells and protein degradation pathways |
Creative Proteomics' Y2H Service Workflow
Project Assessment: Consultation with our experts to understand your research needs and determine the best approach for your project.
Library Construction: Construction of high-quality prey libraries using RNA extraction, cDNA synthesis, and plasmid generation.
Screening: High-throughput screening using our advanced Y2H platform, analyzing protein interactions within a cellular context.
Data Analysis & Interpretation: Our team of experts will process and interpret the data, providing a comprehensive report that includes all identified interactions.
Validation and Confirmation: Additional validation of key interactions through reverse bait-prey assays, co-immunoprecipitation (Co-IP), or pull-down assays.

Why Choose Creative Proteomics for Y2H Services?
Creative Proteomics offers tailored Y2H services with advanced technology, expert team, and high-quality libraries, ensuring reliable results to support your research. Our service advantages:
Custom Library Construction: We provide high-quality cDNA library construction, ensuring diverse coverage from RNA extraction to plasmid creation, enabling detection of a broad range of PPIs.
Accurate In Vivo Interaction Validation: Our service validates interactions in the cellular environment, ensuring biologically meaningful, reliable results.
Detection of Weak and Transient Interactions: Y2H's sensitivity makes it ideal for detecting low-affinity or transient protein interactions often missed by other methods.
Transparent and Affordable Pricing: Our pricing structure is straightforward with no hidden fees, offering great value for comprehensive high-throughput screening.
Comprehensive Data Quality: Our AI-powered platform integrates Y2H data, providing detailed, high-quality reports with insightful analysis.
Features of Y2H Service at Creative Proteomics
Y2H system detectable through growth assays, colorimetric assays, or fluorescence-based systems, providing researchers with both qualitative and quantitative insights into protein interactions.
High Sensitivity: Detects weak or transient interactions that are often missed by other methods.
High Throughput: Enables large-scale screening, saving time and resources while ensuring robust results.
In Vivo Context: Performs interactions within a living yeast cell, providing biologically relevant data.
Comprehensive Library Support: Access to diverse prey libraries enables exploration of complex interaction networks.
Applications of Y2H Analysis
Protein-Protein Interaction Mapping: Identify and confirm interactions between known proteins to create robust interaction networks.
Signaling Pathway Exploration: Study signaling pathways and identify key protein complexes involved in cellular processes.
Functional Gene Discovery: Screen for potential interaction partners of novel genes to reveal their functions.
Mutational Analysis: Perform mutagenesis to pinpoint interaction domains and further dissect the mechanisms behind protein function.
Simple Requirement for Y2H Analysis
| Requirement | Details |
| Volume and Concentration | 1-5 mg/mL; Minimum of 100 µL of each sample |
| Purity | > 80% purity |
| Buffer | Use PBS or appropriate buffer for protein samples; avoid high salt or detergents |
| Storage | Store at -80°C before submission |
| Labeling and Documentation | Clearly label samples with species, sample name, concentration, and date of preparation |
| Shipping | Ship with dry ice to maintain low temperatures |
FAQ
-
Q1: What specific proteins or protein families can be studied using the Y2H system?
A1: Y2H can study various proteins, including transcription factors, enzymes, receptors, structural proteins, and signaling molecules, as long as they are expressed in yeast. Ensuring proper expression of both proteins in yeast cells is crucial. For hard-to-express or membrane-bound proteins, optimizations or alternative approaches may be needed.
-
Q2: Can Y2H be used for high-throughput screening of large protein libraries?
A2: Yes, Y2H is especially useful for high-throughput screening. Our platform is designed to handle large-scale interactions, enabling you to screen complex protein libraries efficiently. We can provide comprehensive cDNA libraries or construct custom libraries to fit specific research goals. Our advanced screening platform allows rapid processing of thousands of potential interactions, ensuring that you get comprehensive data in a timely manner.
-
Q3: What types of reporter systems are available, and how are they read or detected?
A3: Creative Proteomics offers a variety of reporter gene systems, including GFP (Green Fluorescent Protein), lacZ (β-galactosidase), and others. These reporters are chosen based on your experimental needs, and their expression is typically detectable via fluorescence or colorimetric assays. GFP allows for easy visualization under a fluorescence microscope, while lacZ offers a straightforward colorimetric assay through X-gal staining. These reporter systems provide clear, quantifiable readouts of interaction strength and specificity.
-
Q4: How do you handle protein libraries and cDNA construction?
A4: Creative Proteomics uses a high-quality, well-validated process for constructing protein libraries. We begin with RNA extraction from your target tissues or cells, followed by cDNA synthesis, and then create a plasmid library that includes a wide diversity of potential protein interactions. Our optimized protocols ensure that the library is comprehensive and functional, enabling you to identify a broad spectrum of interactions. Custom libraries can also be designed if you need a specific focus or if you want to use specialized protein sources.
-
Q5: What validation methods do you offer to confirm the interactions found through Y2H?
A5: After initial Y2H screening, we offer several validation methods to confirm the interactions detected. These include:
-
Q6: Can you handle protein interactions in difficult or unconventional conditions (e.g., membrane proteins, post-translational modifications)?
A6: Yes, Y2H is commonly used for soluble proteins but can also study membrane-bound proteins and proteins with post-translational modifications (PTMs) with some optimizations. For membrane proteins, we use protocols to enhance expression and stability in yeast. Custom modifications can be added to account for PTMs critical to the interactions being studied, ensuring biologically relevant analysis.
Reverse Bait-Prey Testing: Swapping the bait and prey proteins to confirm interaction specificity.
Co-Immunoprecipitation (Co-IP): This orthogonal technique confirms interactions in a different biological context (in mammalian or yeast cells).
Pull-down Assays: Another confirmation method that works well for validating protein-protein interactions.
These techniques ensure that the interactions detected by Y2H are biologically relevant and not spurious.
Demo
Demo 1: Protein–protein interactions in two potyviruses using the yeast two-hybrid system
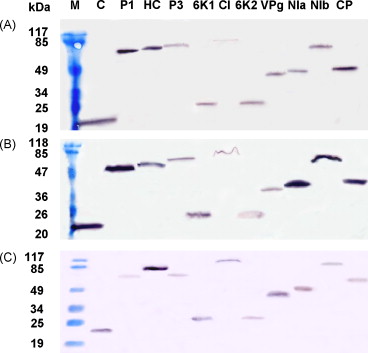
Figure 2. Western blot analysis confirming the expression of the AD and BD fusion proteins from SMV-P and SYSV-O in yeast strain AH109. (A) AD fusions of SYSV-O proteins detected with HA-Tag polyclonal antibody. (B) BD fusions of SYSV-O proteins with c-Myc monoclonal antibody. (C) BD fusions of SMV-P proteins with c-Myc monoclonal antibody. (Lin Lin, et al., 2009)
Demo 2: A two-hybrid system reveals previously uncharacterized protein–protein interactions within the Helicobacter pylori NIF iron–sulfur maturation system
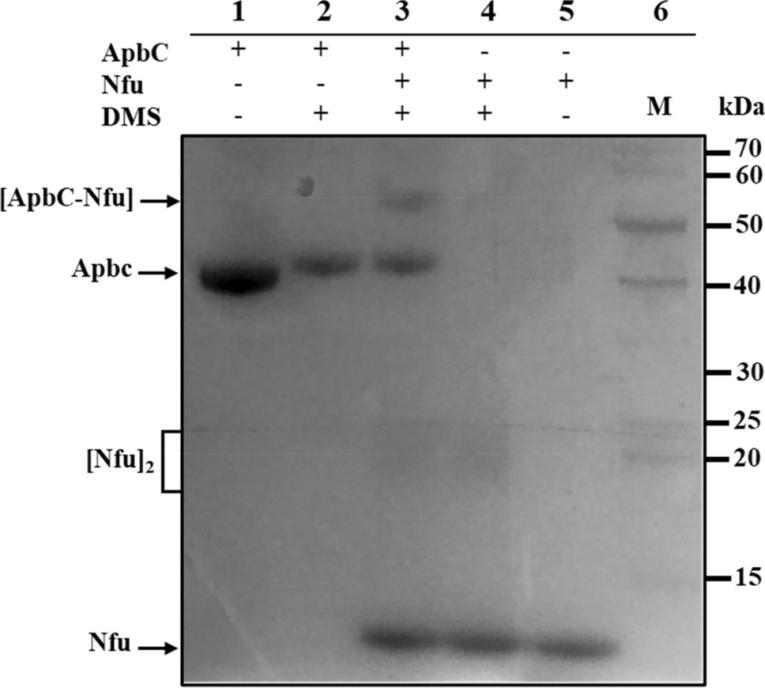
Figure 3. Picture of a gel showing interactions between recombinant ApbC and Nfu proteins. (Stéphane L. Benoit, et al., 2009)
-
Case Study
Case: Using yeast two-hybrid system and molecular dynamics simulation to detect venom protein-protein interactions
Abstract
Snake venom is a complex mixture of proteins and peptides that have various toxic effects during envenomation. Despite the availability of venom proteome and transcriptome data for several snake species, the protein-protein interactions (PPIs) within venom remain poorly understood. These interactions are believed to contribute to the synergistic toxicity of venom, which has critical implications for developing antivenoms. In this study, the authors explore the PPIs within the venom of the Western diamondback rattlesnake (Crotalus atrox) by investigating interactions between phospholipase A2s (PLA2s), a major venom component, and other common venom proteins using a yeast two-hybrid (Y2H) system and molecular dynamics (MD) simulations.
Methods
The study utilized a two-pronged approach combining experimental and computational techniques:
- Yeast Two-Hybrid (Y2H) System: This was used to screen for PPIs between PLA2s and 14 other venom proteins, including Asp49 PLA2, Lys49 PLA2, cysteine-rich secretory protein (CRISP), and others.
- Molecular Dynamics (MD) Simulations: For deeper insights into the structural aspects of identified protein interactions, MD simulations were performed using GROMACS software. The 3D structures of Lys49 PLA2 and CRISP were modeled using MODELLER based on known crystallographic structures and then subjected to docking and dynamic simulations to predict binding modes and assess stability.
Results
- PPIs: The Y2H analysis revealed that PLA2s interact with themselves, confirming previous findings of homodimers (e.g., Lys49 PLA2 homodimers). Additionally, Lys49 PLA2 was found to interact strongly with venom CRISP, while Asp49 PLA2 showed weak interactions with a three-finger toxin (3FTx).
- Molecular Dynamics Simulations: The 3D models of Lys49 PLA2 and CRISP were generated and validated. The most probable binding modes of Lys49 PLA2-CRISP interaction were predicted through protein-protein docking using three different software tools (ClusPro, ZDOCK, HADDOCK). The complex stability was further confirmed through MD simulations, showing that the Lys49 PLA2-CRISP interaction was stable under physiological conditions.
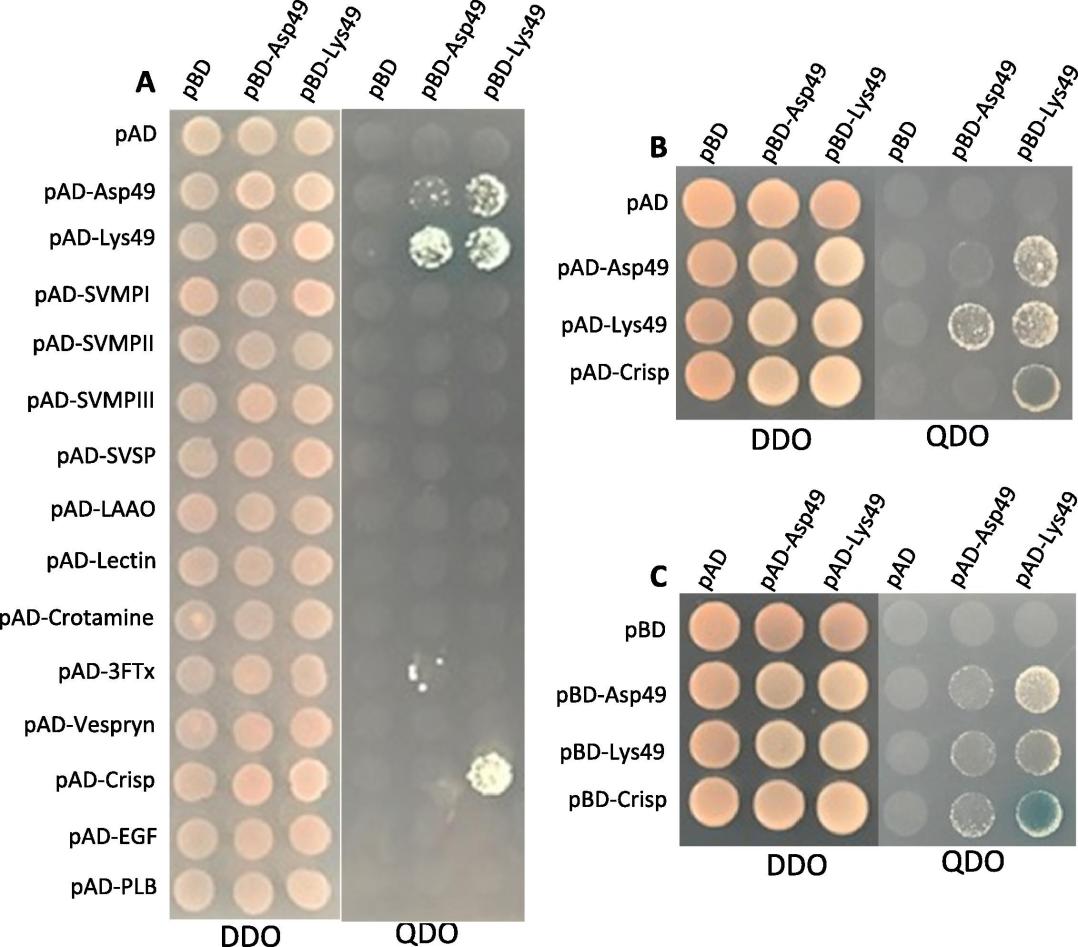
Figure 4. Yeast two-hybrid analysis. Colony growth on DDO medium (without leucine and tryptophan) indicates the successful binary co-transformations, while colony growth on QDO medium (without histidine, leucine, tryptophan, and adenine) shows the PPIs. A, using pBD-Asp49 PLA2 and pBD-Lys49 PLA2 as "bait" to detect the venom PPIs between PLA2s and 14 venom proteins in pAD vector (empty pBD and pAD served as negative control). B, C, the results of 'reciprocal' Y2H, swapping venom proteins from pBD (B) to pAD vector (C), show that PLA2s form dimers and Lys49 PLA2 interacts with CRISP on QDO/X-alpha-Gal medium.
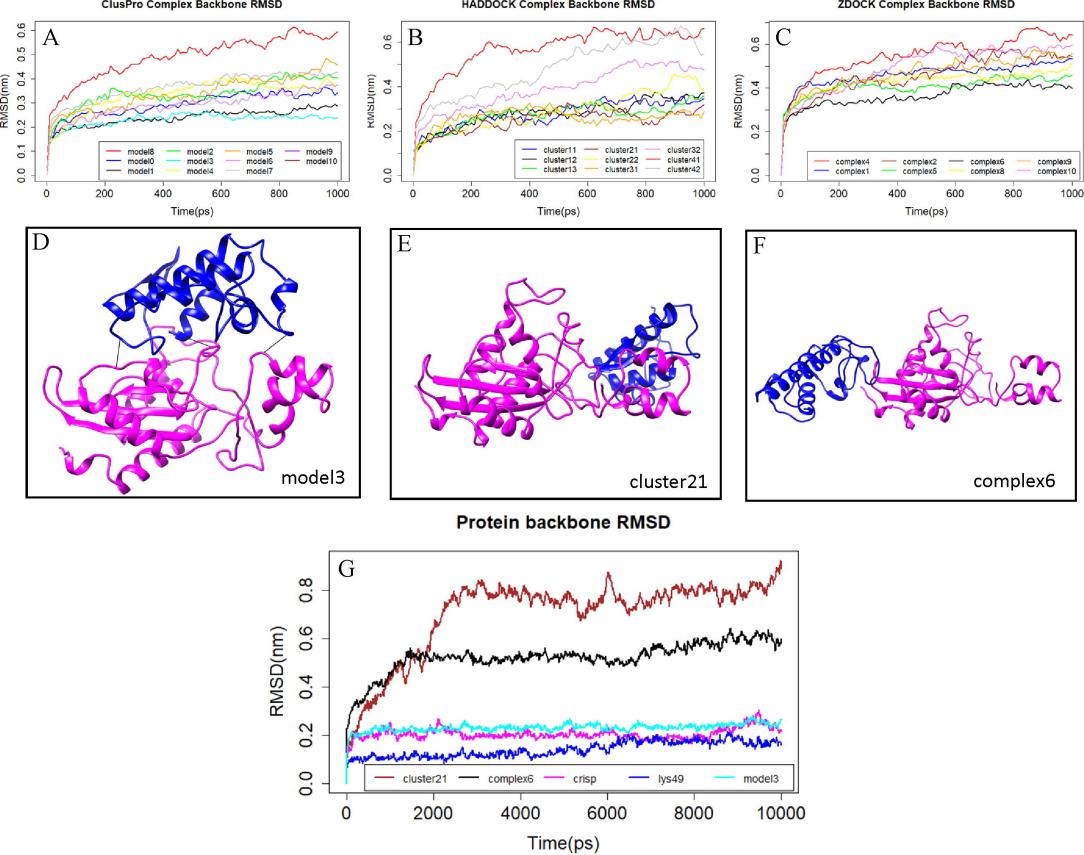
Figure 5. Prediction of Lys49 PLA2-CRISP complex structure by molecular dynamics (MD) simulations. Room-mean-square deviation (RMSD) of backbone atoms from docked modes of (Lys49-CRISP) as a function of simulation time.
Related Services
References
- Jia Y, Kowalski P, Lopez I. Using yeast two-hybrid system and molecular dynamics simulation to detect venom protein-protein interactions. Current Research in Toxicology, 2021, 2: 93-98. DOI: 10.1016/j.crtox.2021.02.006.
- Lechuga A, et al. Unraveling Protein Interactions between the Temperate Virus Bam35 and Its Bacillus Host Using an Integrative Yeast Two Hybrid–High Throughput Sequencing Approach. International Journal of Molecular Sciences, 2021, 22(20): 11105. DOI: 10.3390/ijms222011105
- Benoit S L, Agudelo S, Maier R J. A two-hybrid system reveals previously uncharacterized protein–protein interactions within the Helicobacter pylori NIF iron–sulfur maturation system. Scientific Reports, 2021, 11(1): 10794. DOI: 10.3390/ijms222011105
- Coates P J, Hall P A. The yeast two‐hybrid system for identifying protein–protein interactions. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland, 2003, 199(1): 4-7. DOI: 10.1002/path.1267

