High-Density Protein Microarrays Service
What is High-Density Protein Microarrays?
A High-Density Protein Microarray is a planar substrate bearing a spatially ordered ensemble of protein probes. Each probe occupies a defined feature on the surface. The probes may be full-length proteins, protein fragments, peptides, or in situ expressed products. Arrays permit parallel assays with minimal sample consumption. Readouts commonly use labelled secondary reagents, proximity assays, or enzymatic reporters. The output is high-content, quantitative information on binding, modification, or catalytic activity.
High-Density Protein Microarrays constitute a powerful platform for systematic proteomic interrogation. These platforms enable simultaneous measurement of thousands of protein-analyte interactions. The technology addresses increasing demands for multiplexed profiling in translational research, pharmaceutical discovery, and contract research organizations.
Why Choose High-Density Microarrays for Proteomics
Maximizing Throughput: How high-density arrays enable simultaneous analysis of thousands of proteins, reducing experimental time.
Minimal Sample Requirement: The advantage of low sample consumption for studies with limited biological material.
Broad Proteome Coverage: Comprehensive profiling of full-length proteins, fragments, or peptides to explore diverse protein targets.
Retention of Protein Structure: Importance of correctly folded proteins and post-translational modifications for accurate functional and binding studies.
Quantitative and Reproducible Data: Techniques to reduce background noise, improve signal, and generate reliable, comparable measurements.
Versatility in Applications: Use in antibody-binding studies, functional screens, kinase assays, and small-molecule interactions.
Advanced Technologies for High-Density Protein Microarrays
Content Generation
Recombinant protein production is the primary method for creating array content. Proteins can be made in bacteria, yeast, insect, mammalian cells, or cell-free systems. Each system balances protein yield, correct folding, and chemical modifications differently. Libraries of protein fragments target linear regions, making production easier. Full-length proteins preserve their natural 3D shape and functional sites.
Tagging and Oriented Immobilization
Affinity tags help control how proteins attach to a surface and simplify their purification. Tags like biotin-binding motifs or folding-sensitive reporters allow proteins to bind in a consistent orientation. This alignment keeps active sites and structural regions accessible, ensuring the protein remains functional. In contrast, random chemical attachment can cause proteins to stick in uneven positions, which may reduce their activity.
Surface Chemistry and Microenvironment
Surface coatings, such as polymers and hydrogels, control unwanted binding and help maintain protein stability. Hydrophilic polymers keep proteins in a hydrated environment, preserving their natural shape. Nitrocellulose and epoxide surfaces allow a high density of proteins to be attached. Surfaces that reduce the exposure of water-repelling regions prevent false antibody signals. The surface type also affects how long arrays can be stored, the conditions needed for storage, and the detection sensitivity.
Detection Modalities
Fluorescence scanning remains a principal detection method. Chemiluminescence and label-free optical techniques imaging are applicable for kinetic analysis. Mass spectrometry can verify identity and PTMs. Choice of detection impacts limit of detection, dynamic range, and quantitative linearity.
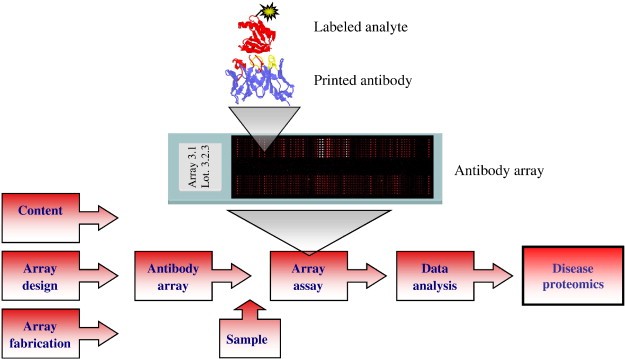
Figure 1. Schematic illustration of the protein microarray set-up (Borrebaeck C A K, et al., 2009).
Creative Proteomics' High-Density Protein Microarrays Service Workflow
- Library Preparation and Expression: Selected ORFs, fragments, or peptide sequences are produced using an appropriate expression system. Quality controls include SDS-PAGE, concentration measurement, and identity verification by mass spectrometry when required.
- Array Fabrication: Purified probes are arrayed under controlled humidity and temperature. Multiple replicates per probe are printed to support statistical analysis. Positive and negative controls are included across the slide.
- Assay Execution: Samples are processed in parallel with standardized incubation times and wash regimens. Detection reagents are validated to avoid cross-reactivity. Internal spike-ins and calibration controls are included to monitor assay performance.
- Image Acquisition and Raw Data Extraction: Slides are scanned with high-resolution imagers. Raw pixel data are extracted for each spot. Saturated pixels are flagged. Neighboring background metrics are computed.
- Data Processing and Statistical Analysis: Background subtraction, replicate consolidation, and probe filtering are executed. CV thresholds identify outlier spots. Differential analysis employs FDR correction.
- Orthogonal Validation: Top candidates proceed to secondary verification. Creative Proteomics offers bead-based multiplex validation, targeted proteomics by parallel reaction monitoring, and conventional immunoassays as orthogonal approaches.

Comparison with Traditional Methods
| Feature | High-Density Protein Microarrays | Traditional Methods (ELISA, Western blot) |
| Throughput | Very High (thousands of proteins per array) | Low (1–10 proteins per assay) |
| Sample Consumption | Low (µL-scale per assay) | High |
| Multiplexing Capability | Extensive | Limited |
| Assay Time | Shorter (parallel analysis in hours) | Longer (days per target) |
| Reproducibility | High, standardized fabrication and replicates | Variable, operator-dependent |
| Functional Assays | Multiple assay types (binding, PTM, enzyme activity) | Possible but limited |
| Discovery vs Targeted Use | Primarily discovery, guides targeted follow-up | Primarily targeted |
| Data Output | Quantitative, high-content | Semi-quantitative |
Applications of High-Density Protein Microarrays
Biomarker Discovery and Cohort Profiling: Arrays enable broad surveys to identify serological markers associated with disease, exposure, or treatment. They facilitate discovery across large antigen repertoires.
Autoantibody and Immunoprofiling Studies: Arrays map immunoglobulin reactivity patterns. They support epitope mapping and cohort stratification.
Antibody Characterization and Epitope Mapping: Arrays aid in assessing antibody specificity, cross-reactivity, and epitope localization. They support antibody engineering and quality control.
Enzyme Substrate Discovery and PTM Mapping: Arrays permit identification of enzymatic substrates. On-array modification assays detect phosphorylation, acetylation, and other post-translational events.
Small-Molecule and Biologic Screening: Arrays enable profiling of compound binding across protein families. The platform supports early selectivity assessments.
Exposome and Environmental Interaction Studies: Arrays quantify humoral responses to environmental antigens. They aid research into exposure-related immune signatures.
Sample Requirements
| Sample Type | Minimum Volume / Amount | Concentration Requirement | Notes |
| Human serum / plasma | 20–50 µL per sample | — | Collect under standardized conditions; avoid repeated freeze–thaw cycles. |
| Antibody isolates | ≥3 µg per sample | >0.1 mg/mL | Purified IgG or monoclonal antibodies preferred; provide buffer details. |
| Cerebrospinal fluid (CSF) | ≥1.0–1.5 mL per sample | — | Clear, cell-free supernatant required; avoid hemolysis. |
| Protein / peptide / small molecule | ≥3 µg per sample | — | Provide information on purification method and solvent composition. |
| Cell / tissue lysates | Equivalent to ≥100 µg total protein | >1 µg/µL | Use validated lysis buffers compatible with protein binding assays. |
| Other biological fluids (e.g., urine, saliva, BALF) | Please inquire | Case-dependent | Contact us for feasibility and optimized protocols. |
Why Choose Creative Proteomics for High-Density Protein Microarrays Service
- Technical Expertise: The scientific staff brings extensive experience in proteomics, assay development, and data analysis. The team leverages robust SOPs and validated reagents.
- Customization and Flexibility: Creative Proteomics offers custom content design, bespoke assay formats, and scalable throughput. The laboratory adapts expression systems and surface chemistries to project needs.
- Quality Assurance and Reproducibility: The company maintains stringent QC checkpoints. Inter- and intra-array variability is monitored. The reporting package supports reproducibility and auditability.
- Client Support and Scientific Consultation: Dedicated project managers and scientific liaisons guide study design and interpretation. The team assists with experimental planning, power calculations, and regulatory alignment.
FAQ
-
Q1: Can High-Density Protein Microarrays measure enzymatic activity directly?
A1: Yes. Arrays can be adapted to detect kinase, phosphatase, acetyltransferase, and ubiquitin ligase activity. Substrates immobilized on the array can be modified enzymatically in situ, with modifications detected using antibodies, fluorescence tags, or mass spectrometry.
-
Q2: How do High-Density Protein Microarrays integrate with mass spectrometry-based proteomics?
A2: Arrays often serve as a discovery platform, with hits validated or quantified using targeted mass spectrometry (PRM, SRM, or DIA). Mass spectrometry provides orthogonal confirmation of protein identity, post-translational modification, and abundance.
-
Q3: Are High-Density Protein Microarrays suitable for post-translational modification (PTM) studies?
A3: Yes. Arrays can be probed with enzymes (e.g., kinases, acetyltransferases) or PTM-specific antibodies to detect phosphorylation, acetylation, methylation, ubiquitination, and SUMOylation. Limitations include incomplete representation of natural PTMs on recombinant proteins, but functional assays on-array provide valuable PTM substrate information.
Demo
Demo 1: Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays
This study used high-density Arabidopsis protein microarrays to systematically identify targets of calmodulin (CaM) and calmodulin-like (CML) proteins, key calcium sensors in plants. Highlighting protein microarrays as a robust platform for high-throughput mapping of plant protein–protein interactions.
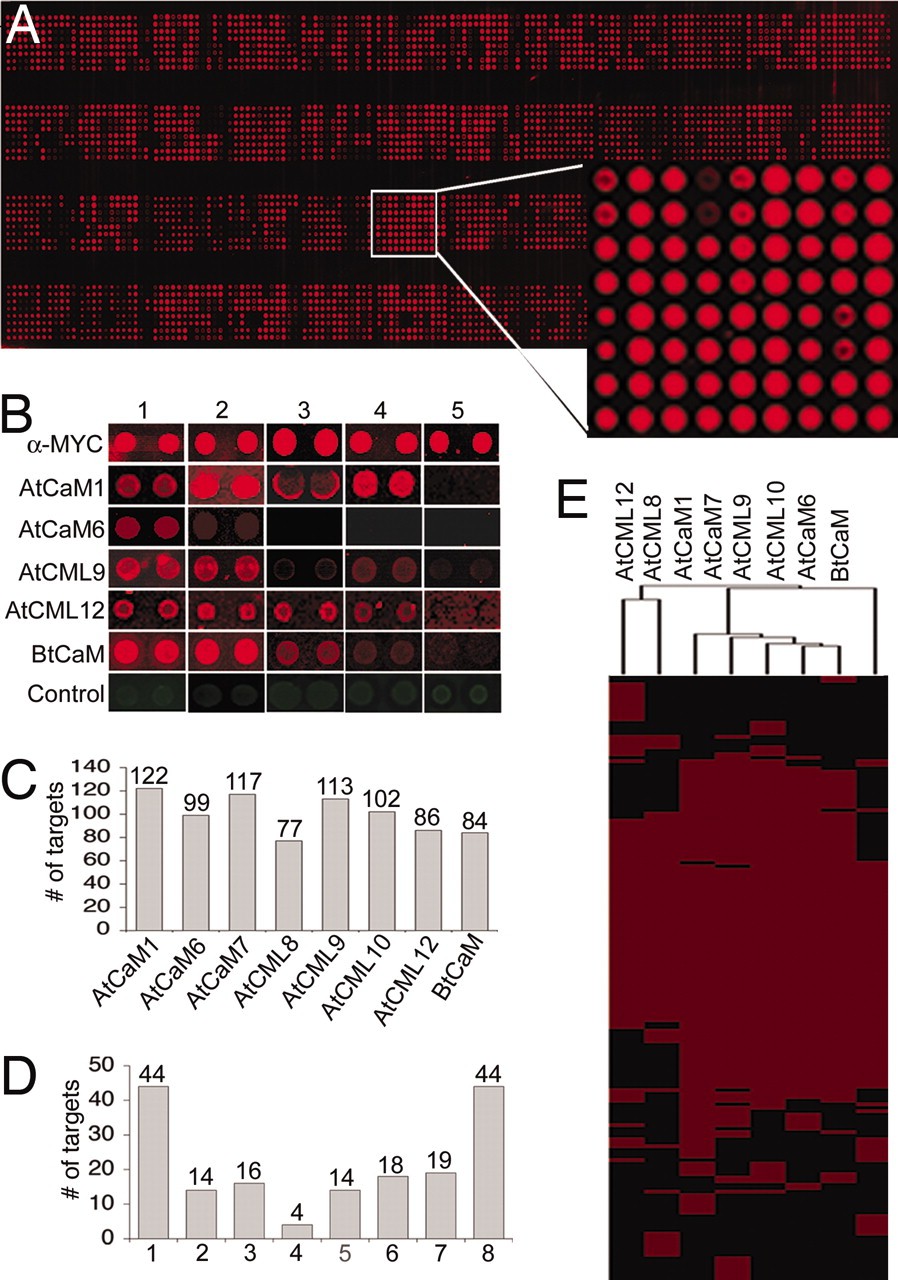
Figure 2. Characterization of the putative CaM/CML substrates on the Arabidopsis protein microarrays (Popescu S C, et al., 2007).
Demo 2: Identification of Tumor-associated Autoantigens for the Diagnosis of Colorectal Cancer in Serum Using High Density Protein Microarrays
This study used high-density protein microarrays containing 8,000 human proteins to identify autoantibody signatures and tumor-associated antigens (TAAs) in colorectal cancer (CRC). Highlighting the potential of these autoantibodies as non-invasive biomarkers for CRC detection and as targets for therapeutic intervention.
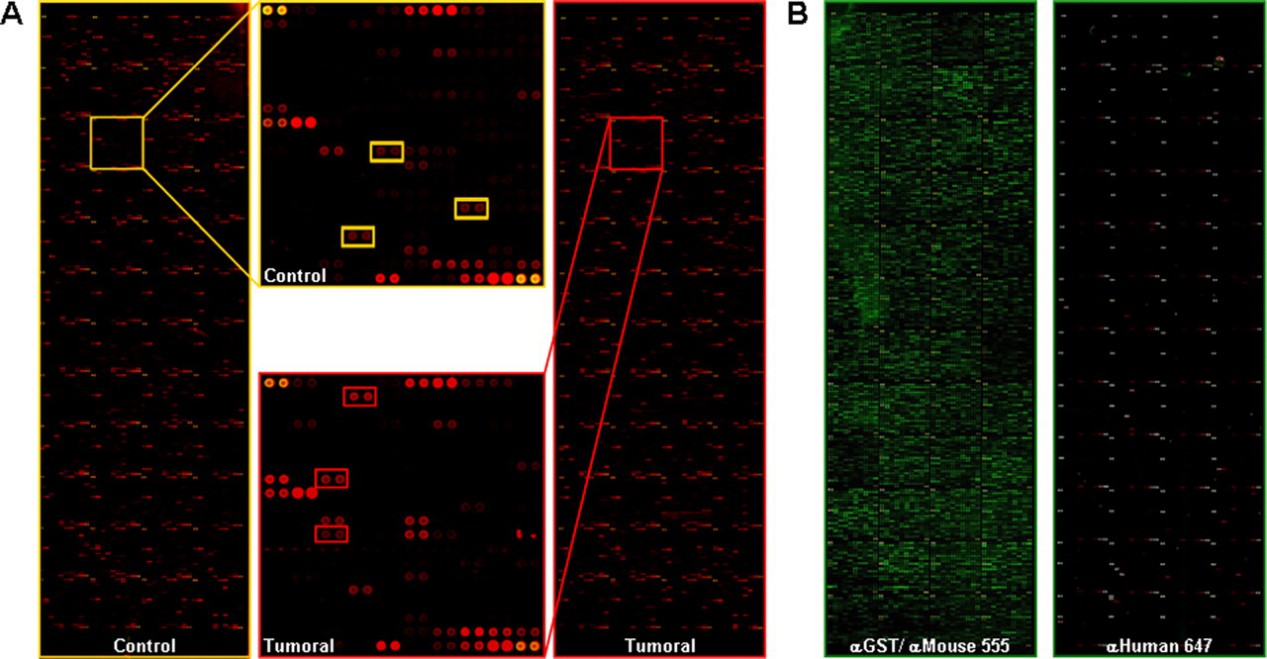
Figure 3. Identification of proteins reactive to autoantibodies (Babel I, et al., 2009).
-
Case Study
Case: Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays
Abstract
Ovarian cancer is a leading cause of cancer-related deaths in women, with poor survival rates for late-stage disease. Early detection is critical, but existing markers, such as CA-125, are insufficiently sensitive for early-stage diagnosis. Identifying proteins differentially expressed in ovarian cancer could improve diagnostics and reveal therapeutic targets. The study aimed to identify ovarian cancer-associated proteins using high-density protein microarrays probed with patient sera, to discover tumor-associated autoantigens that are overexpressed in cancer tissue relative to healthy tissue.
Methods
Sera from 30 ovarian cancer patients and 30 healthy individuals were screened against high-density human protein microarrays containing 5,005 proteins. Autoantibody binding was detected using fluorescent secondary antibodies. Candidate antigens were identified using three statistical approaches: paired t-tests, ReliefF feature selection, and Proteome Prospector software. Selected proteins were validated using dot blot, Western blot, and immunohistochemistry on tissue microarrays.
Results
- Ninety-four proteins exhibited enhanced reactivity in cancer patient sera.
- Four candidates—lamin A/C, SSRP1, RALBP1, and ZNF265—were further analyzed. Lamin A/C, SSRP1, and RALBP1 showed elevated expression in ovarian cancer tissue relative to healthy tissue; ZNF265 did not.
- Lamin A/C and SSRP1 staining outperformed CA-125 in sensitivity (95%) and specificity (97.5%).
- These markers were also present in other cancer types, reflecting broader tumor-related protein dysregulation.
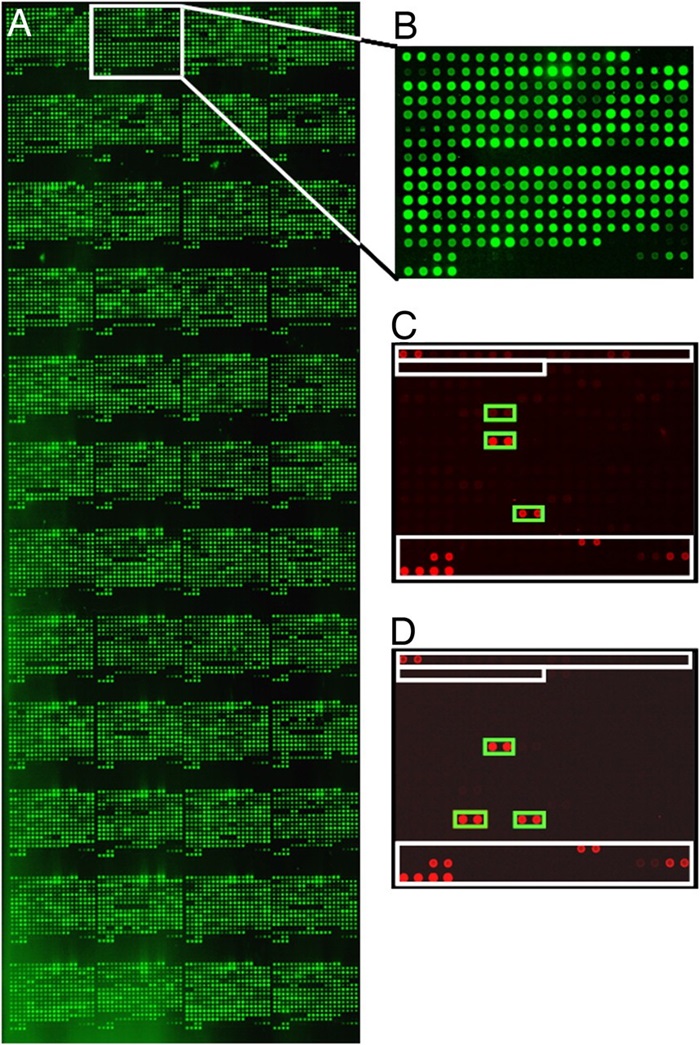
Figure 5. Identification of tumor-associated autoantibodies and targeted protein antigens.
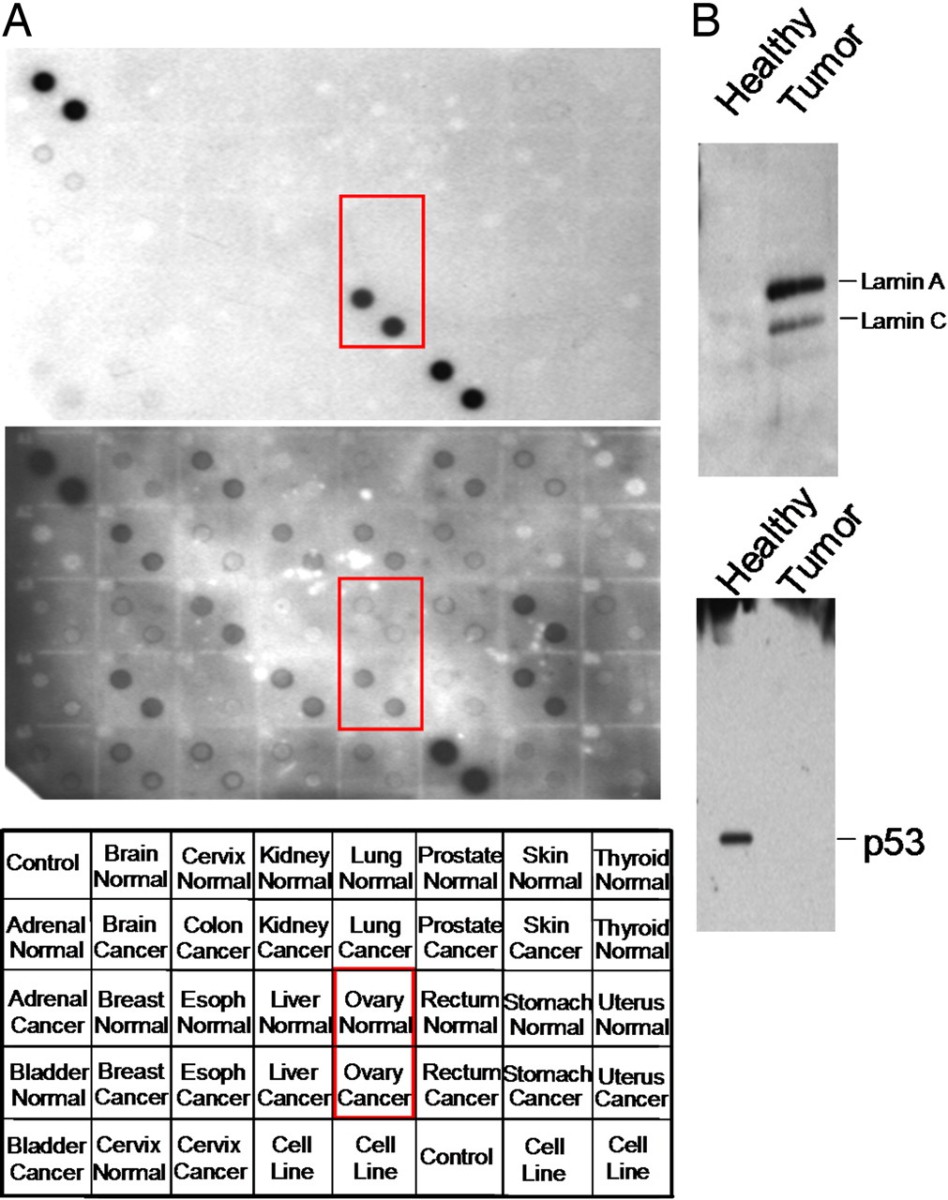
Figure 6. Immunoblot analysis of candidate tumor markers.
Conclusion
High-density protein microarrays effectively identified candidate tissue biomarkers for ovarian cancer, including lamin A/C and SSRP1, which exhibited robust signatures in tissue and outperformed conventional markers like CA-125. This approach demonstrates the power of protein-level screening for biomarker discovery. It highlights the potential of microarrays to reveal disease-related protein changes that are not detectable by RNA profiling alone.
Related Services
References
- Borrebaeck C A K, Wingren C. Design of high-density antibody microarrays for disease proteomics: key technological issues. Journal of proteomics, 2009, 72(6): 928-935.
- Popescu S C, et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proceedings of the National Academy of Sciences, 2007, 104(11): 4730-4735.
- Babel I, et al. Identification of tumor-associated autoantigens for the diagnosis of colorectal cancer in serum using high density protein microarrays. Molecular & cellular proteomics, 2009, 8(10): 2382-2395.
- Hudson M E, et al. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proceedings of the National Academy of Sciences, 2007, 104(44): 17494-17499.