HUMAN Quantitative Proteomics Arrays
What are Quantitative Proteomics Arrays?
Quantitative proteomics arrays are advanced tools designed to measure multiple proteins simultaneously, providing a comprehensive view of the protein composition in cells, tissues, or body fluids. Unlike traditional single-protein tests, these arrays can simultaneously analyze hundreds or even thousands of proteins, revealing patterns and interactions that help researchers understand complex biological systems. By combining multiple non-overlapping sections, an array can cover up to 1,000 human proteins, including enzymes, signaling molecules, and immune regulators. These platforms are compatible with a wide variety of samples, including blood, serum, tissue extracts, and cell culture fluids, making them highly flexible for various research applications. Their quantitative output enables researchers to track changes in protein levels, compare healthy and diseased states, or assess the effects of experimental treatments, all in a high-throughput, efficient, and reproducible manner.
Advantages of Using Quantitative Proteomics Arrays
Quantitative proteomics arrays offer several key advantages over traditional protein detection methods such as single-plex ELISA or western blotting:
- High Throughput and Coverage: With the ability to measure thousands of proteins simultaneously, these arrays reduce sample consumption while providing comprehensive profiling of the proteome.
- Quantitative Accuracy: Standardized detection reagents and controlled assay conditions allow for precise, reproducible quantification across multiple experiments.
- Versatility: The arrays are compatible with a wide range of sample types, enabling studies of human tissue, blood, cell lysates, and other biological fluids.
- Time Efficiency: Multiplexing significantly shortens experimental timelines compared to serial single-protein measurements, supporting rapid biomarker screening and hypothesis testing.
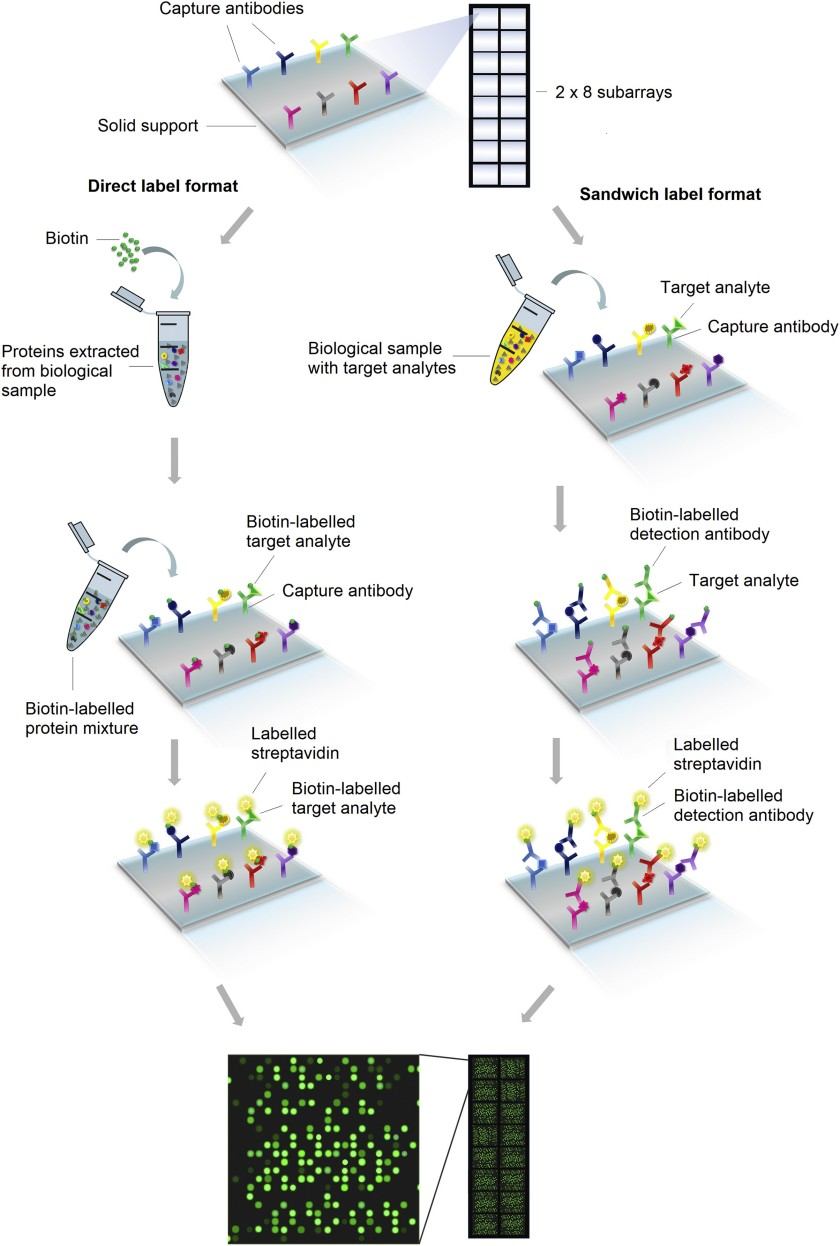
Figure 1. Workflow of planar proteomics microarray technologies (Ren A H, et al., 2021).
Comparison with Alternative Proteomics Approaches
- Throughput: Arrays provide simultaneous measurement of hundreds to thousands of proteins, while ELISA typically measures one protein per assay.
- Sample Requirements: Arrays require lower sample volumes than serial single-plex assays, preserving precious samples.
- Coverage: Arrays offer broad proteome coverage, whereas mass spectrometry may require complex sample preparation and data analysis for equivalent protein identification.
- Ease of Use: Immunoassay-based arrays offer a straightforward workflow with standardized reagents, reducing technical variability and training requirements.
Creative Proteomics' HUMAN Quantitative Proteomics Arrays Service Workflow
- Sample Preparation: Biological samples are clarified, diluted, and prepared under controlled conditions.
- Array Blocking: Non-specific binding sites on the array surface are blocked to minimize background noise.
- Sample Incubation: Prepared samples are applied to the array, allowing target proteins to bind to their corresponding capture antibodies.
- Detection Antibody Incubation: Biotinylated or labeled secondary antibodies are applied to the array to detect bound proteins.
- Signal Detection: Fluorescence or chemiluminescence signals are captured using compatible imaging systems.
- Data Analysis: Fluorescence intensity is quantified and normalized, often using software tools to calculate relative protein concentrations and generate heatmaps or statistical analyses.

Data Analysis and Interpretation
- Normalization: Adjusting for sample variability and background signals to ensure accurate comparisons across arrays.
- Reproducibility: Performing replicate measurements to confirm protein quantification consistency.
- Statistical Analysis: Using appropriate bioinformatics tools to identify significantly altered proteins or patterns associated with experimental conditions.
- Visualization: Generating heatmaps, clustering, and network diagrams to interpret complex proteomic data effectively.
Applications of HUMAN Quantitative Proteomics Arrays
- Biomarker Discovery and Validation: Researchers can identify candidate biomarkers for diseases, monitor therapeutic responses, and validate protein targets identified through other omics approaches.
- Pathway and Network Analysis: By simultaneously quantifying multiple proteins involved in cellular signaling or metabolic pathways, the arrays facilitate systems biology studies and the mapping of complex molecular interactions.
- Drug Discovery and Preclinical Research: Quantitative profiling of protein expression in cellular models or tissue samples helps evaluate target engagement, off-target effects, and pharmacodynamic responses in experimental therapeutics.
- Immune Profiling: Comprehensive measurement of cytokines, chemokines, and immune regulatory proteins enables investigation of immune system dynamics, inflammation, and host responses.
- Translational Research: Integration of proteomic data with genomic, transcriptomic, or metabolomic datasets allows researchers to explore disease mechanisms and identify potential translational biomarkers for future clinical applications.
Choosing the Right Quantitative Array for Your Project
Selecting the appropriate quantitative proteomics array is a critical step to ensure reliable, actionable results for any biomarker discovery or proteome profiling study. You must consider several key factors when choosing an array.
- Type and quality of the biological sample: such as plasma, serum, tissue lysates, or cell culture supernatants—can influence the choice of array, as some platforms are optimized for specific sample matrices to maximize sensitivity and reduce background noise.
- Biomarker coverage and number of targets: For instance, a study focused on immune signaling may require an array enriched for cytokines and chemokines, whereas a broader disease profiling study may benefit from a high-coverage array detecting hundreds to thousands of proteins.
- Sensitivity and dynamic range: Particularly when target proteins are present at low concentrations, as arrays with higher signal-to-noise ratios provide more reliable quantification.
- Throughput requirements: It can determine whether a single-slide kit or a multi-slide high-throughput format is more suitable.
Sample Requirements
| Sample Type | Minimum Volume | Preparation Notes |
| Serum / Plasma | ~50–100 µL | Collect using standard protocols; avoid repeated freeze–thaw cycles. |
| Cell Culture Supernatant | ~100–200 µL | Clarify by centrifugation to remove debris before application. |
| Tissue Lysates | 50–100 µg protein | Homogenize tissues under cold conditions; normalize protein concentration. |
| Cell Lysates | 50–100 µg protein | Lyse under non-denaturing conditions; quantify total protein before use. |
| Other Body Fluids | ~50–100 µL | Pre-clear samples as necessary to reduce particulate matter and viscosity. |
Why Choose Creative Proteomics for HUMAN Quantitative Proteomics Arrays
- Comprehensive Target Coverage: 1,000 biomarkers across multiple biological pathways.
- Expert Support: Guidance from experienced proteomics specialists with decades of industry expertise.
- Flexible Service Models: Options for kit-only, scanning, or full-service analysis.
- Fast Turnaround: Results delivered with minimal downtime to keep projects on track.
FAQ
-
Q1: What is the dynamic range of detection for these arrays?
A1: The arrays offer a wide dynamic range, allowing for the detection of both high-abundance and low-abundance proteins, which is essential for comprehensive proteomic analysis.
-
Q2: Can these arrays be used for longitudinal studies?
A2: Yes, these arrays are suitable for longitudinal studies, allowing researchers to monitor changes in protein expression over time, which is valuable for tracking disease progression or treatment effects.
-
Q3: Can these arrays detect protein isoforms?
A3: Detection of protein isoforms depends on the specificity of the antibodies used; some arrays may include antibodies that can distinguish between isoforms.
-
Q4: Can array data be integrated with proteomics from mass spectrometry or transcriptomics?
A4: Yes. Arrays measure targeted protein abundance via antibodies, while MS provides broader, often unbiased coverage. Integrative analysis should account for detection biases, dynamic range differences, and post-transcriptional regulation when comparing to transcriptomics.
Demo
Demo: Investigating a novel multiplex proteomics technology for detection of changes in serum protein concentrations that may correlate to tumor burden.
In a pilot translational study, the authors applied a 1,000-protein quantitative antibody microarray to pre- and post-surgical serum from pancreatic cancer patients to identify proteins that decreased after tumor resection. The array identified several putative hits, but orthogonal validation with single-target immunoassays showed limited replication for many candidates — the paper emphasizes that high-throughput antibody arrays can generate hypotheses but require stringent orthogonal validation to avoid false positives. The study is useful as an example of array application in small patient sets and of the critical importance of follow-up validation.
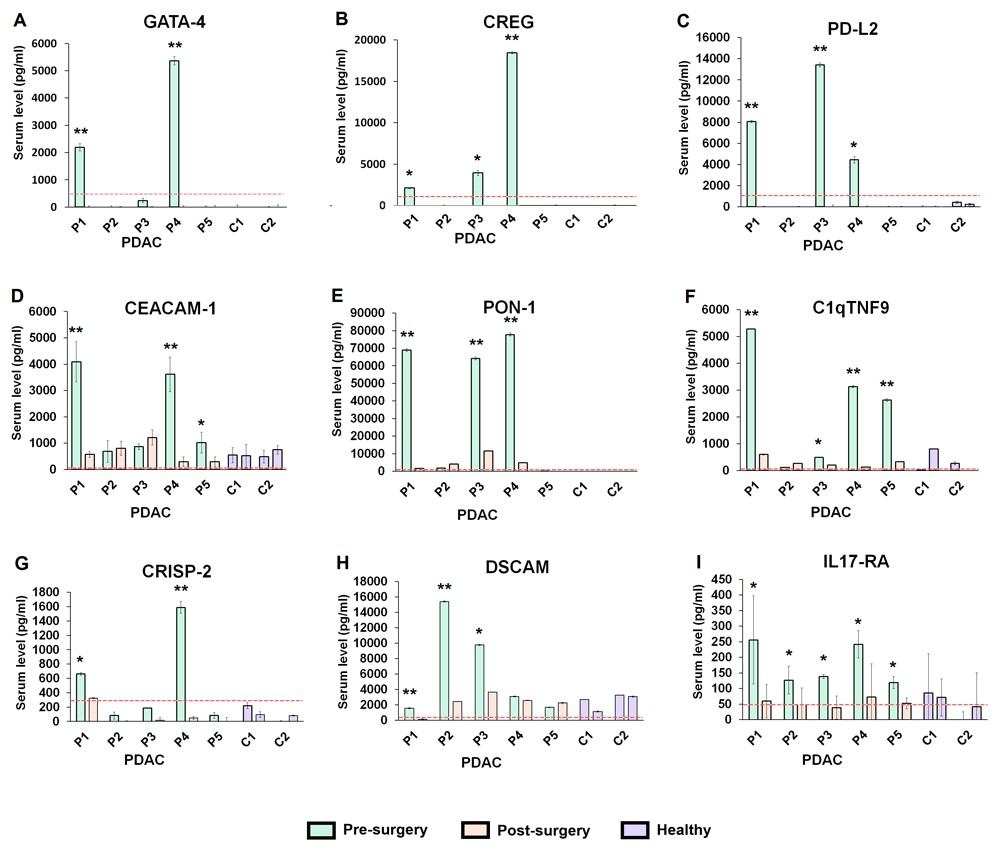
Figure 2. Multiplexed array results for the proteins that decreased in serum level following surgery in pancreatic cancer (PDAC) patients (Ren A H, et al., 2020).
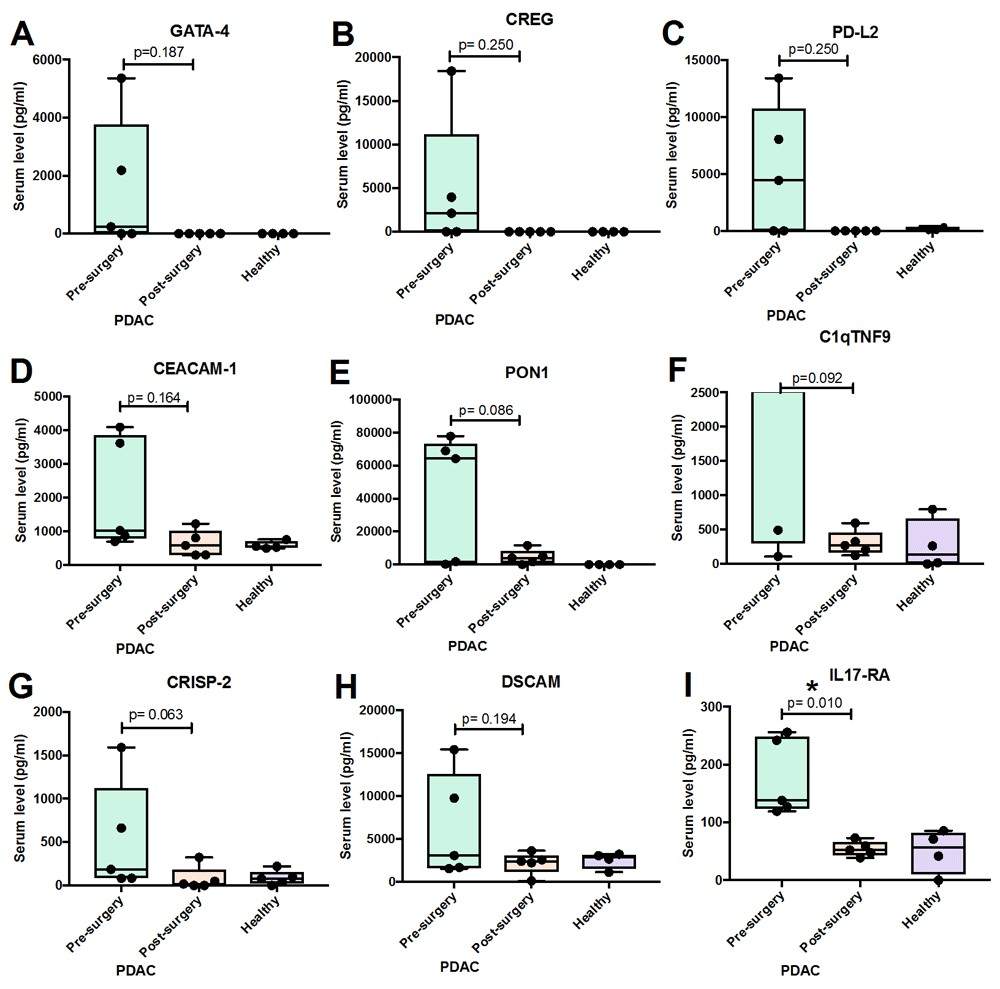
Figure 3. Group analysis of proteins that showed decrease in serum level in individual pancreatic cancer (PDAC) patients collected before and after surgery (Ren A H, et al., 2020).
-
Case Study
Case: Novel CSF biomarkers for diagnosis and integrated analysis of neuropsychiatric systemic lupus erythematosus: based on antibody profiling
Abstract
Neuropsychiatric systemic lupus erythematosus (NPSLE) is difficult to diagnose due to heterogeneous symptoms and the lack of reliable biomarkers. Cerebrospinal fluid (CSF), which directly reflects brain pathology, may provide more specific diagnostic information than blood-based markers. The study aimed to identify and validate novel CSF protein biomarkers for the diagnosis of NPSLE and to explore their potential link with disease mechanisms.
Methods
- Discovery phase: Antibody microarray screening of ~1000 proteins in CSF from controls, SLE, and NPSLE patients.
- Candidate selection: Protein–protein interaction analysis and literature review identified 17 candidates.
- Validation phase: Custom 17-plex antibody microarray tested CSF from larger cohorts (controls, SLE, NPSLE).
- Analysis: Differential expression, logistic regression, ROC curves, and integration with hippocampal transcriptomics from a lupus mouse model.
Results
- 29 differentially expressed proteins were identified in discovery screening.
- Five validated biomarkers—CST6, TCN2, KLK5, L-selectin, and Trappin-2—showed significant diagnostic value.
- Combined biomarker panels achieved high diagnostic accuracy: CST6 + TCN2 + KLK5 (AUC = 0.880) for NPSLE vs. SLE. CST6 + L-selectin + Trappin-2 + TCN2 (AUC = 0.908) for NPSLE vs. controls.
- Correlations with disease activity and overlap with lupus mouse hippocampal data supported biological relevance.
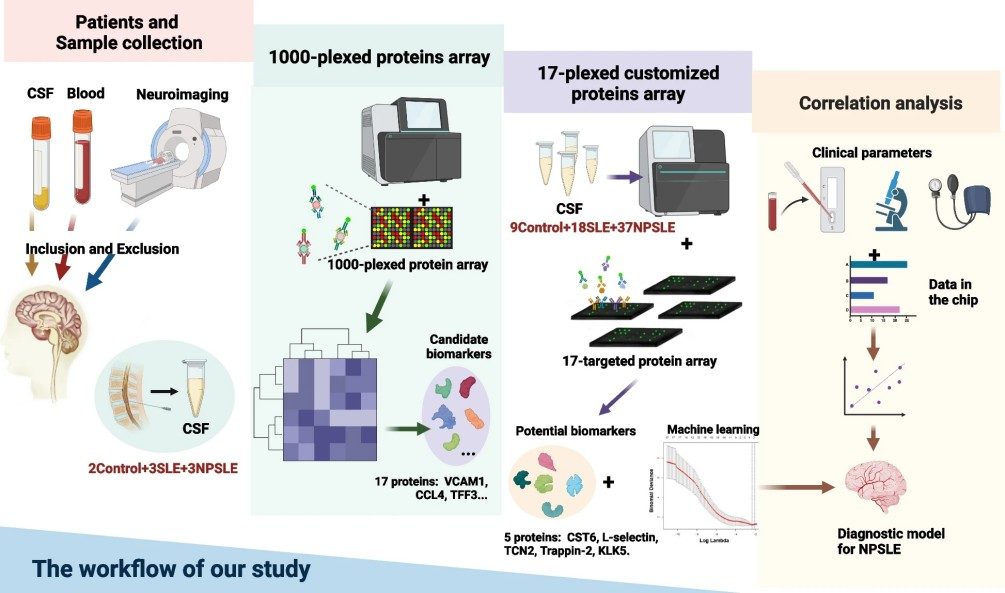
Figure 4. Workflow of one thousand CSF proteins array screening.
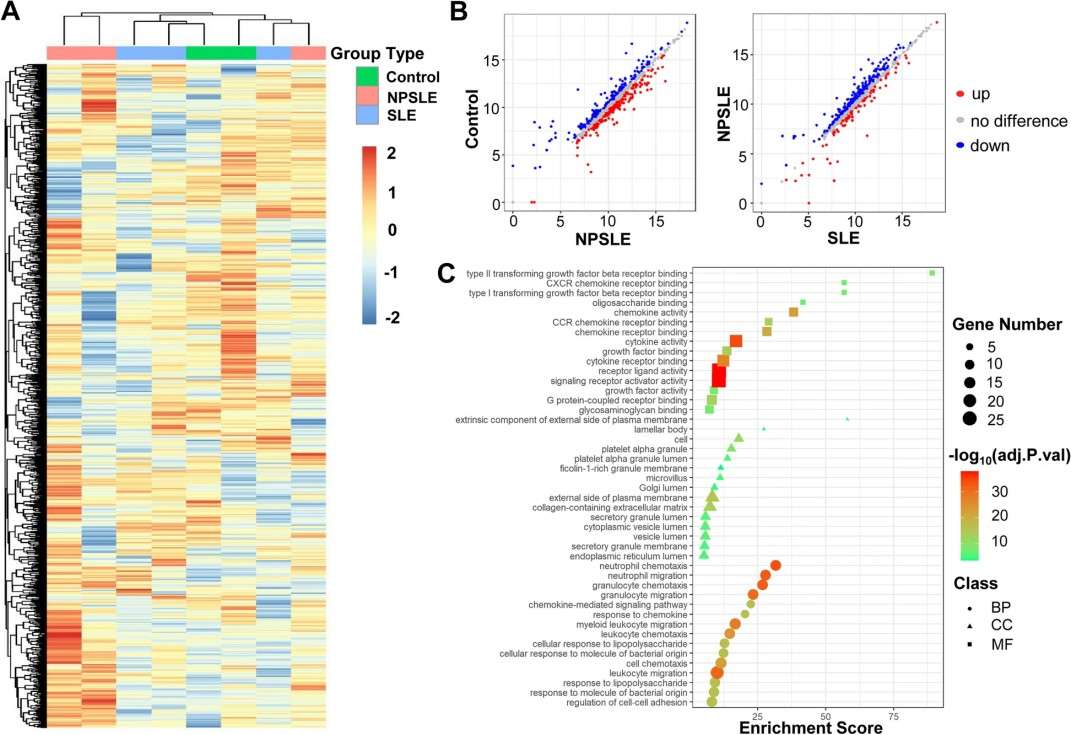
Figure 5. One thousand-plexed proteins array of CSF samples.
Conclusion
A panel of CSF protein biomarkers was identified and validated for NPSLE diagnosis. These findings suggest that multiplex CSF proteomic profiling can improve diagnostic accuracy and provide insights into disease mechanisms, particularly those related to neuroinflammation and cognitive dysfunction.
Related Services
References
- Ren A H, et al. Investigating a novel multiplex proteomics technology for detection of changes in serum protein concentrations that may correlate to tumor burden. F1000Research, 2020, 9: 732.
- Ren A H, Diamandis E P, Kulasingam V.Uncovering the depths of the human proteome: antibody-based technologies for ultrasensitive multiplexed protein detection and quantification. Molecular & Cellular Proteomics, 2021, 20: 100155.
- Ni J, et al.Novel CSF biomarkers for diagnosis and integrated analysis of neuropsychiatric systemic lupus erythematosus: based on antibody profiling. Arthritis Research & Therapy, 2023, 25(1): 165.