Multiplex ELISA Arrays Services
Accelerating Biomarker Discovery through Multiplex ELISA Arrays?
Multiplex ELISA arrays represent a highly advanced platform for the simultaneous quantification of multiple proteins within a single sample. This approach integrates the specificity and sensitivity of traditional enzyme-linked immunosorbent assays with the high-throughput capabilities of microarray technology. The platform allows researchers to measure up to forty analytes concurrently, conserving precious sample material and increasing experimental efficiency. Multiplex ELISA arrays are particularly suitable for biomarker discovery, cytokine profiling, disease mechanism studies, and pharmacodynamic investigations. This service is intended exclusively for research purposes.
What Are Multiplex ELISA Arrays and How Do They Work?
Multiplex ELISA arrays are a glass-chip version of the familiar sandwich ELISA that lets you measure many proteins at once from a single small sample. Instead of coating a microwell plate for one protein at a time, this platform prints tiny spots of different capture antibodies onto a glass slide. Each spot is specific for one protein (for example, an individual cytokine), and the spots are arranged in a defined grid so the instrument knows which protein sits where.
Step-by-step
- Apply sample: A diluted biological sample (serum, plasma, cell culture medium, or lysate) is added so proteins can bind to their matching capture spots.
- Wash: Unbound material is removed to reduce background.
- Add detection antibody: A second antibody that recognizes a different part of the same protein is applied. This creates a "sandwich" around the target protein, improving specificity.
- Signal generation: The detection antibody carries a reporter (either fluorescent dye or an enzyme that produces light). The slide is scanned and the intensity at each spot is measured. Brighter spots mean more of that protein.
- Quantification: Known concentration standards run on the same slide produce a calibration curve; sample signal intensities are converted to absolute concentrations from that curve.
Key practical points
- Controls and replicates: Each run includes positive and negative controls and often replicate spots for each analyte to check reproducibility.
- Cross-reactivity control: Antibody pairs are pre-tested to avoid unwanted binding between antibodies and non-target proteins; spatial separation of spots further reduces interference.
- Dynamic range and sensitivity: Performance varies by analyte and depends on antibody affinity and the detection method; some proteins are measured down to low picogram/mL levels, others require higher concentrations.
- Sample needs: The platform is sample-efficient (microliter volumes); however, proper dilution and pre-treatment are important to avoid matrix effects.
Fundamental Principles of Multiplex ELISA
- Capture Antibody Immobilization: Each glass chip or microarray surface is prepared with tiny, precisely arranged spots, where capture antibodies are immobilized. Each spot corresponds to a specific target protein, enabling the simultaneous measurement of multiple proteins within a single sample.
- Target Protein Binding: When the biological sample is applied, proteins naturally seek out and bind to their matching capture antibodies.
- Detection Antibody Formation: After the capture step, a second antibody, as the detection antibody, is introduced. This antibody recognizes a different site on the same protein, creating a "sandwich" around the target. The detection antibody is linked to a reporter system, often fluorescent or chemiluminescent, which enables the generation of a signal.
- Signal Generation and Imaging: The bound detection antibodies generate a measurable signal. Fluorescence scanners or imaging systems capture the intensity of each spot, producing a detailed map of protein expression.
- Specificity and Cross-Reactivity Control: Antibody pairs are carefully selected to avoid unintended binding, known as cross-reactivity. Spatial separation of capture spots further minimizes interference between different targets, which is critical when analyzing complex biological mixtures.
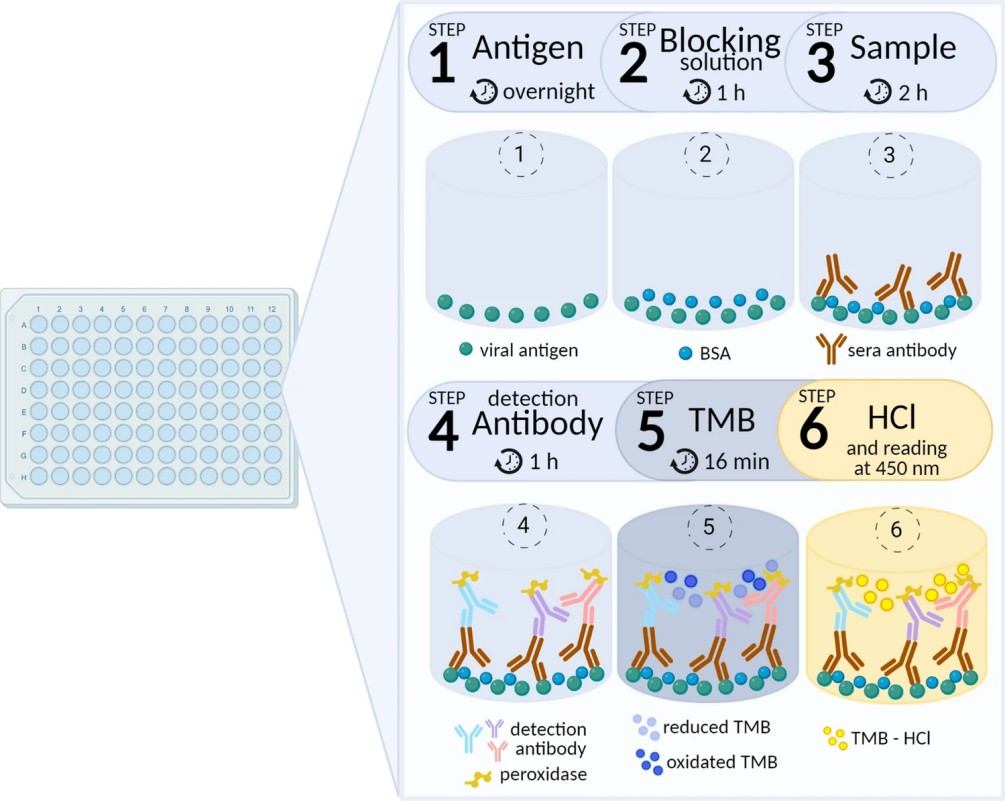
Figure 1. The steps of multiplex ELISA assay (Fernandes-Siqueira L O, et al., 2022).
Comparison: Multiplex ELISA vs. Conventional ELISA
| Attribute | Multiplex ELISA Array | Conventional Single-Analyte ELISA |
| Sample volume | Very low (small microliter volumes per sample). | Higher (tens to hundreds of microliters per analyte). |
| Throughput | Very high when automated; hundreds of samples feasible. | Moderate; throughput limited by plate format and per-analyte setup. |
| Operational cost | Lower cost per analyte when multiplexed at scale. | Higher cost per analyte for multi-analyte studies. |
| Data richness | High; enables profile and signature analysis across pathways. | Limited; focused quantitation of single analytes. |
| Analytical sensitivity | High for many analytes; depends on antibody affinity and detection modality. | High; often optimized for each analyte individually. |
| Precision and reproducibility | High within validated panels; intra-array quadruplicates improve robustness. | High for validated kits; reproducible when protocol and reagents are strictly controlled. |
| Suitability for discovery studies | Very well suited; enables simultaneous hypothesis generation and biomarker screening. | Less suited; best for targeted validation of individual biomarkers. |
| Sample types supported | Serum, plasma, cell culture supernatant, tissue lysate with appropriate pre-treatment. | Serum, plasma, tissue lysate, and others; typically validated per kit. |
Creative Proteomics' Multiplex ELISA Arrays Service Workflow
- Sample Submission and Preparation: Receipt and verification of sample integrity. Optimization of sample dilution and treatment to suit the multiplex array.
- Plate/Chip Preparation: Selection or customization of glass chip-based arrays with specific protein panels. Verification of capture antibody spotting and array quality control.
- Assay Execution: Addition of samples to the subarrays. Incubation under controlled conditions to allow target binding. Application of detection reagents for chemiluminescent or fluorescent signal generation.
- Signal Detection: Imaging of arrays using high-resolution imaging systems. Capture of quantitative signal intensity for each analyte.
- Data Analysis: Background subtraction, normalization, and standard curve fitting. Generation of concentration data for all measured proteins. Optional consultation for interpretation and integration with downstream analyses.
- Quality Control and Reporting: Verification of assay reproducibility and precision across replicates. Delivery of validated, ready-to-use data with detailed reporting for research purposes.

Applications of Multiplex ELISA Arrays in Biomedical Research
- Cytokine and Chemokine Profiling: Quantification of immune mediators to investigate inflammatory responses, immune signaling, and disease pathogenesis.
- Biomarker Discovery: Identification of novel protein markers for disease diagnosis, prognosis, and therapeutic monitoring.
- Drug Mechanism Studies: Evaluation of pharmacodynamic effects and signaling pathway modulation in preclinical and translational research.
- Disease Mechanism Elucidation: Analysis of complex protein networks to understand molecular interactions underlying disease processes.
- Validation of Antibody Arrays: Confirmation and quantification of results obtained from semi-quantitative protein arrays.
- High-Throughput Screening: Efficient profiling of multiple samples for large-scale studies, enabling population-level or multi-condition analyses.
Sample Requirements
| Sample Volume | 50 µL per assay (typical; may vary with panel size) |
| Sample Types | Serum, plasma, cell culture supernatant, tissue lysates |
| Species Supported | Human, mouse, rat, pig (expandable to other species upon request) |
| Number of Samples per Slide | Up to 14 samples per glass chip, depending on standard allocation |
| Number of Proteins per Panel | Up to 40 proteins simultaneously |
Why Choose Creative Proteomics for Multiplex ELISA Arrays Service
- Expertise and Experience: Creative Proteomics has rich experience in proteomics and assay development, ensuring reliable and scientifically robust results.
- High Sensitivity and Accuracy: The platform combines glass chip-based multiplex ELISA with precise detection and quantification, minimizing variability and maximizing reproducibility.
- Customizable Panels: Researchers can select up to 40 proteins per array, tailoring panels to specific study objectives and sample types.
- High-Throughput Capability: The service supports simultaneous processing of multiple samples, enabling large-scale studies with efficient turnaround.
- Comprehensive Support: Creative Proteomics provides full technical assistance, including assay design, sample preparation, data analysis, and interpretation, ensuring seamless integration into research workflows.
FAQ
-
Q1: Can the panel be customized to include specific proteins of interest?
A1: Yes. Panels can be tailored to include up to 40 proteins relevant to the study. Researchers can select combinations of cytokines, chemokines, growth factors, or other biomarkers. Customization ensures that the assay aligns precisely with experimental objectives.
-
Q2: Are control samples provided?
A2: Yes. Each assay run includes positive and negative controls, as well as a set of calibration standards. Controls allow researchers to verify assay performance and detect any technical issues. Q3: Is it possible to track longitudinal changes in protein expression?
-
Q3: Is it possible to track longitudinal changes in protein expression?
A3: Yes. The service supports serial measurements across multiple time points. Researchers can monitor changes in protein levels over time, during treatment, or under different experimental conditions.
-
Q4: What is the maximum number of samples that can be processed in one run?
A4: The number depends on chip layout, panel size, and automation. Typically, multiple chips can be nested in standard trays, allowing dozens of samples to be processed simultaneously.
Demo
Demo: Extensive cytokine biomarker analysis in serum of Guillain-Barré syndrome patients.
Guillain–Barré syndrome (GBS) is an acute immune-mediated polyneuropathy. The syndrome is associated with antecedent infection and dysregulated immune responses. The etiopathogenesis remains incompletely characterized. Serum cytokine and protein signatures may reveal pathophysiologic mediators and candidate biomarkers for diagnosis or prognosis. The authors motivated broad proteomic screening to discover such serum biomarkers.
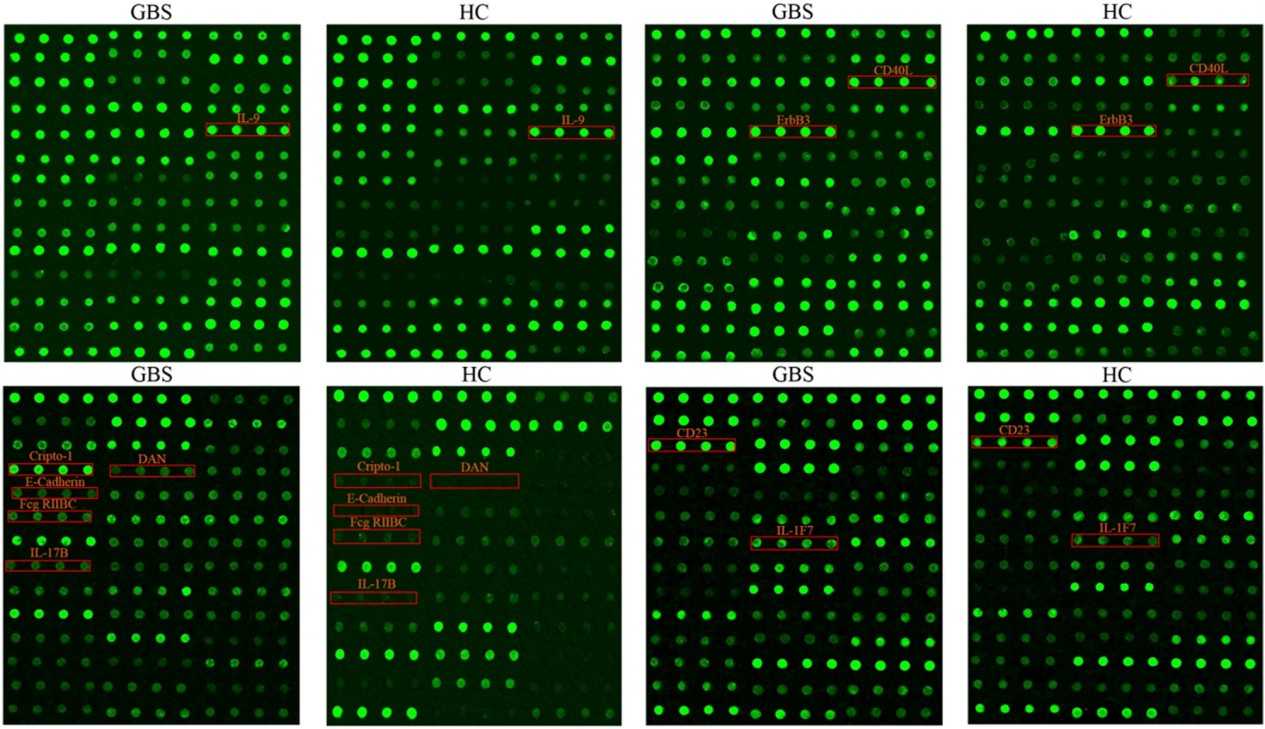
Figure 2. The top 10 DEPs profiles in GBS patients (Li X, et al., 2023).
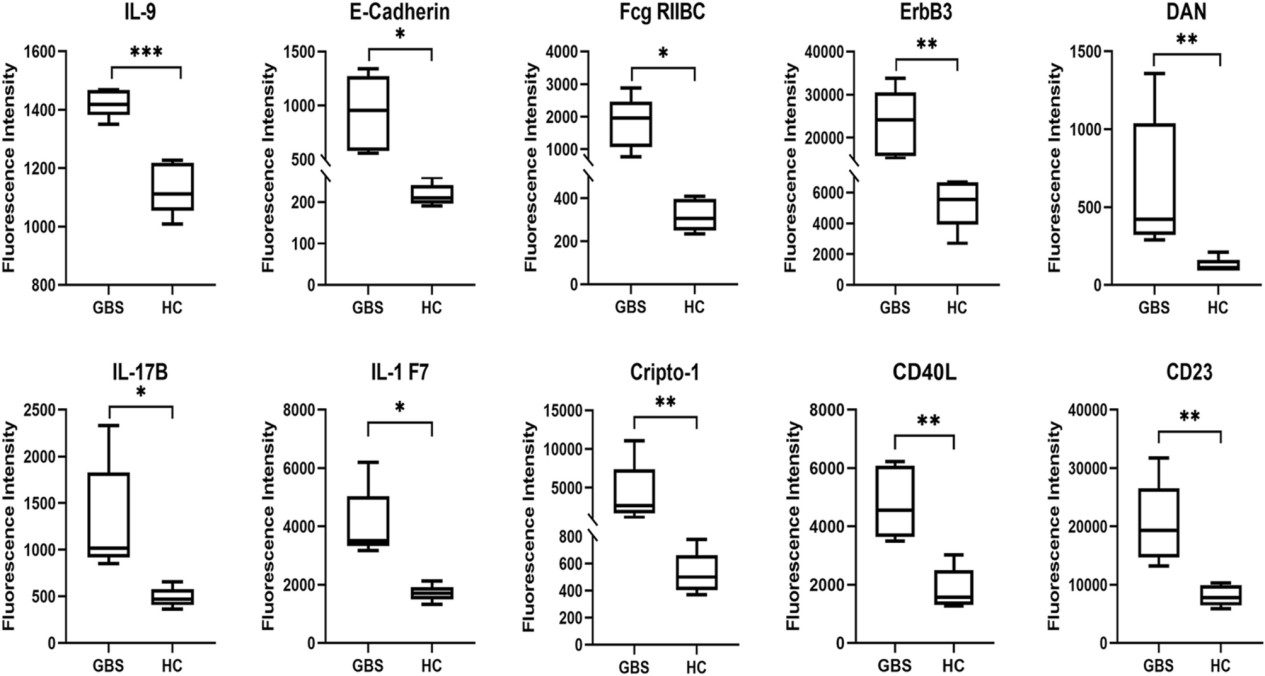
Figure 3. Boxplots were used to analyze the fluorescence intensity of the top 10 DEPs (Li X, et al., 2023).
-
Case Study
Case: Extensive cytokine biomarker analysis in serum of Guillain-Barré syndrome patients.
Abstract
Accurate serological tests are essential for monitoring SARS-CoV-2 infection, immunity, and vaccine responses. Multiplex assays detecting multiple antigens (S, RBD, N-NTD) and antibody isotypes (IgG, IgA, IgM) provide a comprehensive approach. To develop a cost-effective, in-house multiplex ELISA for detecting IgG, IgA, and IgM against key SARS-CoV-2 antigens, suitable for assessing infection- and vaccine-induced seroconversion.
Methods
- Cohorts: 126 individuals (23 PCR-positive, 103 non-diagnosed) in Rio de Janeiro; subset vaccinated with CoronaVac or ChAdOx1 nCoV-19.
- Antigens: Trimeric S, RBD, N-NTD expressed in HEK293T or E. coli.
- Assay: ELISA optimized for antigen coating, serum/detection antibody dilution, and incubation. Heat-inactivated sera; cut-offs defined using pre-pandemic samples and ROC/Youden index analyses.
Results
- PCR + samples: 100% IgG to S, 78% to RBD, 52% to N-NTD; IgA and IgM responses varied.
- Non-diagnosed individuals: Initial low seropositivity (≈12%); seroconversion increased over 5 months.
- Vaccinated individuals: S IgG induced in all; RBD IgG stronger in PCR + subjects; minimal N-NTD or IgM response.
- The multiplex assay efficiently distinguished between infection- and vaccine-induced responses, showing cross-reactivity and variability in N-NTD responses.
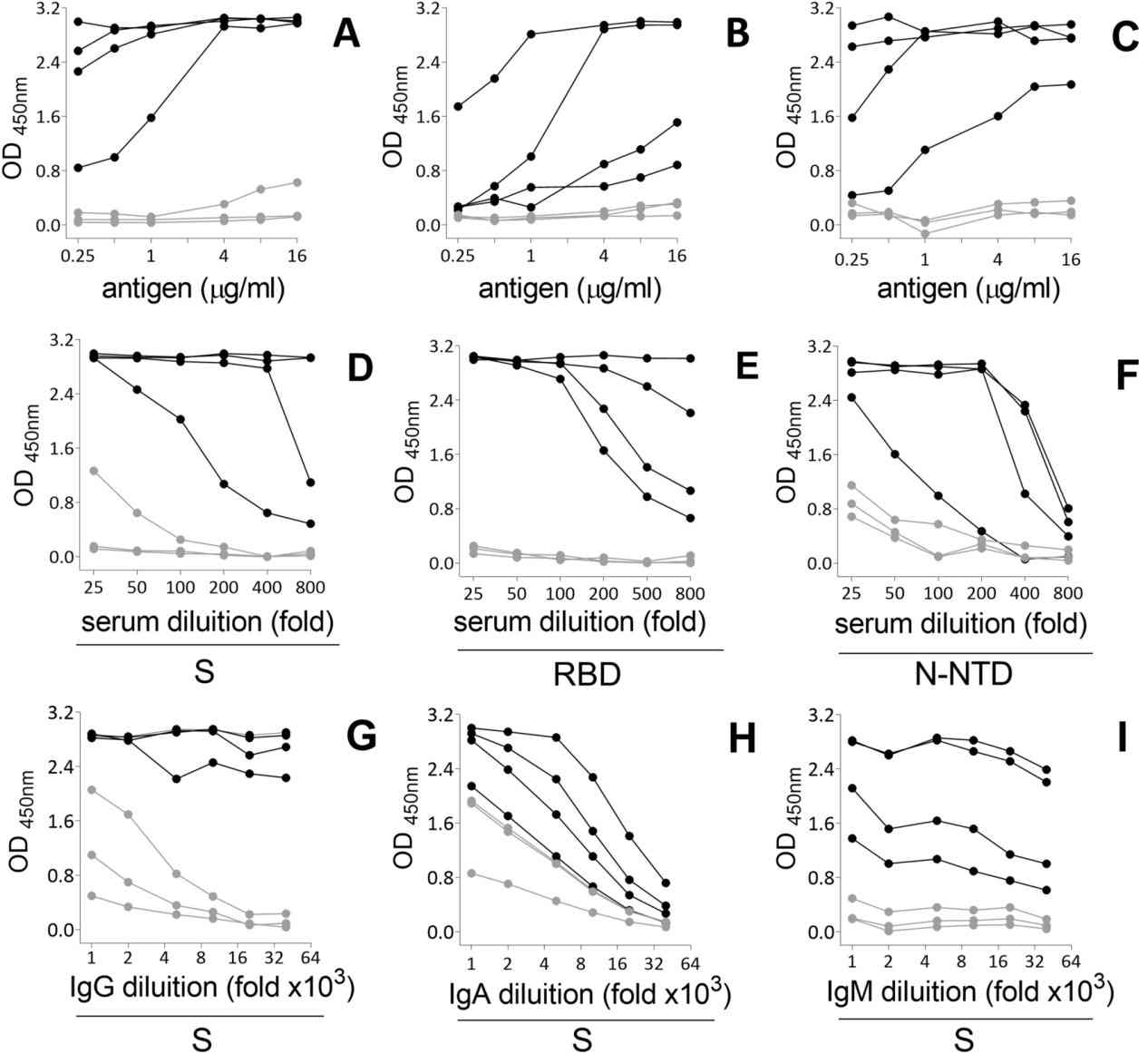
Figure 4. Titration of serum antibodies against SARS-CoV-2 S protein.
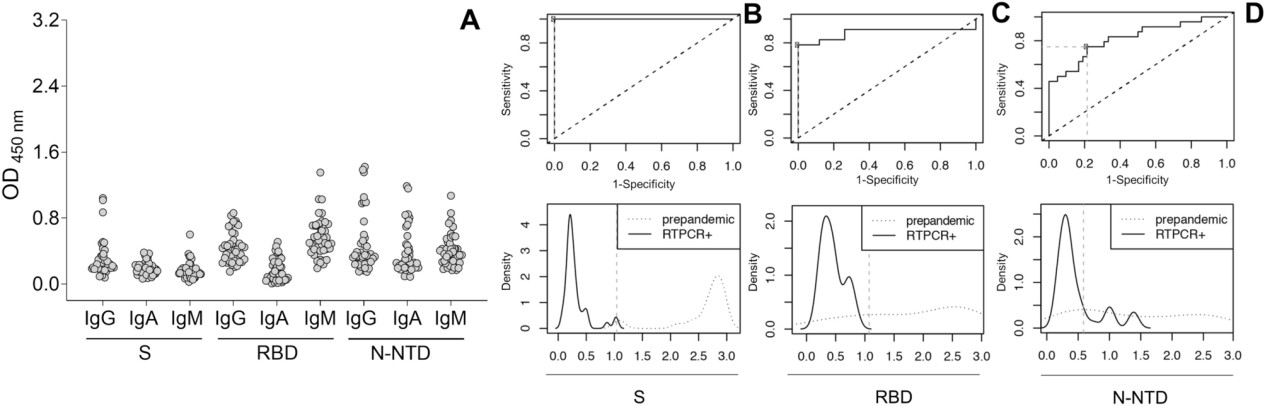
Figure 5. Cut-off determination in the optimized in-house ELISA.
Conclusion
The multiplex ELISA is a flexible, sensitive, and cost-effective tool for monitoring seroconversion and vaccine responses. Its ability to measure multiple antigens and antibody isotypes enhances diagnostic accuracy and informs public health strategies, particularly amid emerging variants. Limitations include reliance on pre-pandemic samples for cut-offs and potential cross-reactivity with other viral proteins.
Related Services
References
- Leng S X, et al. ELISA and multiplex technologies for cytokine measurement in inflammation and aging research. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences, 2008, 63(8): 879-884.
- Li X, et al.Extensive cytokine biomarker analysis in serum of Guillain-Barré syndrome patients. Scientific Reports, 2023, 13(1): 8354.
- Fernandes-Siqueira L O, et al.On the caveats of a multiplex test for SARS-CoV-2 to detect seroconversion after infection or vaccination. Scientific Reports, 2022, 12(1): 10366.