- Services
- FAQ
- Demo
- Case Study
- Related Services
- Support Documents
- Inquiry
What is C-terminal sequence
The C-terminal sequence is an important structural and functional part of proteins and peptides, and plays a role in the biological function of proteins. In the analysis process of recombinant protein expression and purification products, especially the development and process establishment of recombinant protein products, the C-terminal sequence of the protein needs to be identified and analyzed to study the type of degradation products of protein drugs and the specificity of enzyme cleavage. Our protein C-terminal sequencing technology service can provide C-terminal sequencing analysis methods for biopharmaceutical proteins or other biological reagents such as MALDI-ISD for Top-down protein C-terminal sequencing and carboxypeptidase hydrolysis.
Creative Proteomics is a reliable biopharmaceutical partner with professional proteomics research resources. We use first-class experimental platform and mature technical methods to provide you with high-quality one-stop service. The quality control of the biopharmaceutical industry requires information on the identification and structural analysis of the C-terminal sequence of protein and peptide drug molecules. In particular, the ICH Q6B guidelines require the C-terminal sequence information of protein (peptide) drugs. To this end, we will provide you with comprehensive N-terminal sequencing technology services. We will provide you with comprehensive and GLP / cGMP-compliant N-terminal sequencing analysis service around the ICH guidelines (especially ICH Q6B) and the US FDA issues 'Points to Consider' document.
We Can Provide but Not Limited to
- Analysis of amino acid release
- Advanced structural analysis of proteins
- C-terminal sequence analysis
- Target peptide fragmentation poor quality analysis
- C-terminal modification site type analysis
- Protein molecular analysis
Technology Platform of C-terminal Sequencing Analysis Service
Creative Proteomics uses MALDI-ISD to perform Top-down protein sequencing, mass spectrometry analysis technology, carboxypeptidase kinetics technology, C-terminal amino acid release technology and other technologies to effectively and accurately analyze the protein C-terminal sequence.
We provide the following analysis methods:
1) MALDI Top-Down Sequencing
MALDI-ISD is used to break the inter-residue bond of the target peptide, which mainly cleaves the N-Cα bond of the main chain of the target peptide, so that it is fragmented in the mass spectrometer, and then is cleaved by the inter-residue bond by analysis. The mass difference between the generated series of consecutive fragment ions is used to identify the C-terminal sequence of the target peptide. The specific workflow is as follows.
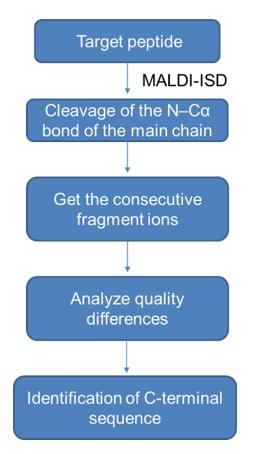
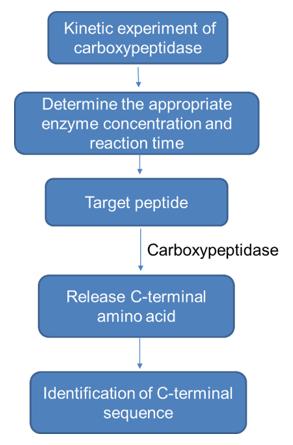
2) Carboxypeptidase hydrolysis method
First, carry out the kinetic experiment of carboxypeptidase to determine the appropriate enzyme concentration and reaction time, so that the released amino acids are mainly C-terminal amino acids. According to the relationship between the amount of released amino acids (moles) and the reaction time, the peptide chain can be identified C-terminal amino acid sequence. The specific workflow is as follows.
Advantages of C-terminal Sequencing Analysis Service
- Short time-consuming: MALDI-ISD for Top-down protein sequencing method does not require proteinase digestion by protease, it can quickly identify the C-terminal sequence of the protein and confirm the type of C-terminal modification, which greatly shortens the time for sample identification and analysis.
- High sensitivity: Using mass spectrometry analysis methods can get high sensitivity during the analysis of C-terminal sequence.
- High-throughput: In this service, the C-terminal sequence of the protein can be combined by MALDI-ISD Top-down protein C-terminal sequencing and carboxypeptidase hydrolysis method, these two methods can complement each other to obtain complete C-terminal sequence analysis information.
- Customized service: We can customize professional solutions for you according to your research plan needs. You can select or suggest the required items for analysis.
Creative Proteomics's professional researchers can provide customers with complete C-terminal sequence analysis information of proteins to speed up your research on advanced protein structure, C-terminal sequence analysis and modification types. We will provide you with detailed experimental design and final C-terminal sequence information analysis report and other data reports.
Sample Requirements
| Sample Format |
|
| Sample Quantity |
|
| Sample Purity |
|
| Storage & Handling |
|
FAQ
Q: How do you handle the potential presence of post-translational modifications (PTMs) in C-terminal sequencing?
A: Post-translational modifications (PTMs) can significantly influence the structure and function of proteins, particularly at the C-terminus. In our C-terminal sequencing services, we take several steps to address the presence of PTMs:
Sample Preparation: We ensure that the sample is processed under conditions that minimize the risk of PTM alteration. This includes maintaining appropriate pH and temperature during enzymatic digestion.
Specific Analytical Techniques: Our use of mass spectrometry is particularly advantageous, as it can detect mass shifts corresponding to various PTMs. For instance, modifications like phosphorylation, methylation, or glycosylation can be identified by observing changes in the mass-to-charge (m/z) ratios of fragment ions.
Detailed Reporting: Our reports provide insights into the types of modifications detected at the C-terminus, helping researchers understand how these changes may affect protein function and stability.
By employing these strategies, we ensure a comprehensive analysis that considers the potential impact of PTMs on the C-terminal sequence.
Q: What is the differences between C-terminal and N-terminal sequencing?
A: C-terminal and N-terminal sequencing are both crucial techniques in proteomics, but they focus on different ends of the protein chain and serve distinct purposes:
C-terminal Sequencing: This process involves identifying the amino acids at the C-terminus of the protein. It is important for understanding protein stability, function, and the presence of modifications at the C-terminus. C-terminal sequencing is particularly relevant in applications like quality control of biopharmaceuticals and the study of proteolytic processing.
N-terminal Sequencing: In contrast, N-terminal sequencing focuses on the amino acids at the N-terminus of the protein. This is often done using Edman degradation and is useful for confirming protein identity, determining protein purity, and analyzing protein modifications. N-terminal sequencing can provide insights into protein synthesis and processing.
Both techniques complement each other and are essential for a comprehensive understanding of protein structure and function. Depending on the research question, either method—or both—may be necessary for a complete analysis.
Demo
Demo: C-terminal Sequence Analysis of Peptides by MALDI-MS After Digestion with Carboxypeptidases.
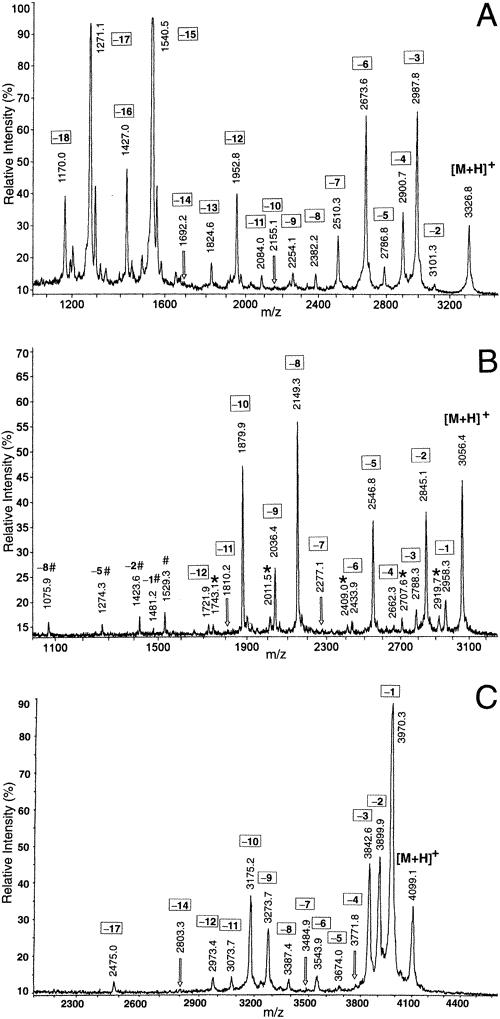 Figure 1. C-terminal sequence analysis of underivatized peptides by MALDI-MS after digestion with vasoactive intestinal polypeptide (A), secretin (B), and fragments of yeast alcohol dehydrogenase (C).
Figure 1. C-terminal sequence analysis of underivatized peptides by MALDI-MS after digestion with vasoactive intestinal polypeptide (A), secretin (B), and fragments of yeast alcohol dehydrogenase (C).Case Study
Case: Improving de Novo Sequencing of Peptides Using a Charged Tag and C-Terminal Digestion
Background
The article discusses a novel strategy for peptide de novo sequencing that improves upon existing methodologies by utilizing TMPP-Ac-OSu (a charge derivatization reagent) and enzymatic digestion. Traditional sequencing methods face challenges with peptide identification, especially in complex biological samples where fragmentation patterns can be ambiguous. The incorporation of charge derivatization enhances the mass spectrometric analysis by promoting the formation of sequence-specific ions and improving the interpretability of tandem mass spectrometry (MS/MS) data. Additionally, the authors note that current algorithms in de novo sequencing software are not optimized for the fragmentation patterns of TMPP-Ac derivatives, suggesting a need for specialized software to enhance accuracy.
Methods
- Peptide Derivatization: Peptides are derivatized using TMPP-Ac-OSu, optimizing conditions to 80% acetonitrile with a 10-fold excess of reagent, achieving higher conversion yields.
- SCX Chromatography: Excess TMPP reagent is removed using strong cation exchange (SCX) chromatography, effectively isolating the derivatized peptides from hydrolyzed products.
- Enzymatic Digestion: Carboxypeptidase B (CPB) digestion is performed post-SCX cleanup to selectively remove C-terminal Arg or Lys residues from the derivatized peptides, with careful control of enzyme/substrate ratios and digestion time.
- Reversed-Phase Liquid Chromatography (RPLC): Modified peptides are separated via RPLC, facilitating their subsequent analysis.
- MALDI-TOF MS Analysis: The eluted peptides are deposited on a MALDI target for analysis. Data are collected in DDA mode, followed by processing to generate peak lists for de novo sequencing software.
Results
The method was evaluated on model proteins (BSA and yeast ADH), leading to the following findings:
- Peptide Sequencing Success: From the analysis, 19 peptides for BSA and 10 for ADH were sequenced. Out of 29 peptides analyzed, 20 achieved exact matches to known sequences.
- Error Analysis: Among the discrepancies, three peptide sequences differed by a single amino acid, attributed to the misidentification of lysine as glutamine due to their minimal mass difference. The remaining six sequences provided partial information, mainly due to the presence of proline residues disrupting the ion series during fragmentation.
- Software Limitations: The de novo sequencing software, while effective, struggled with TMPP-Ac derivatives due to differences in fragmentation patterns compared to traditional protonated peptides. The authors highlight the necessity for improved algorithms tailored for TMPP-Ac fragmentation.
- Calibration and Accuracy: The incorporation of the TMPP tag as an internal calibrant facilitated mass accuracies of 5 ppm or better, enhancing the reliability of the sequencing process.
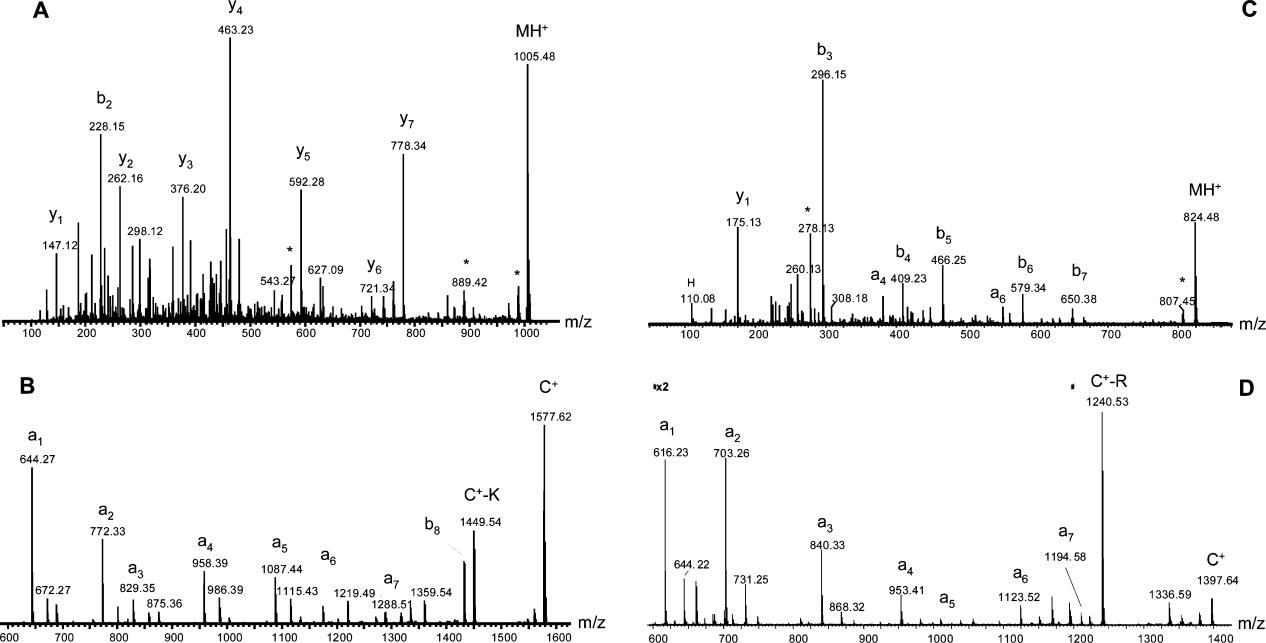 Figure 2. MALDI Q-TOF tandem mass spectra of peptide standards and their derivatives.
Figure 2. MALDI Q-TOF tandem mass spectra of peptide standards and their derivatives.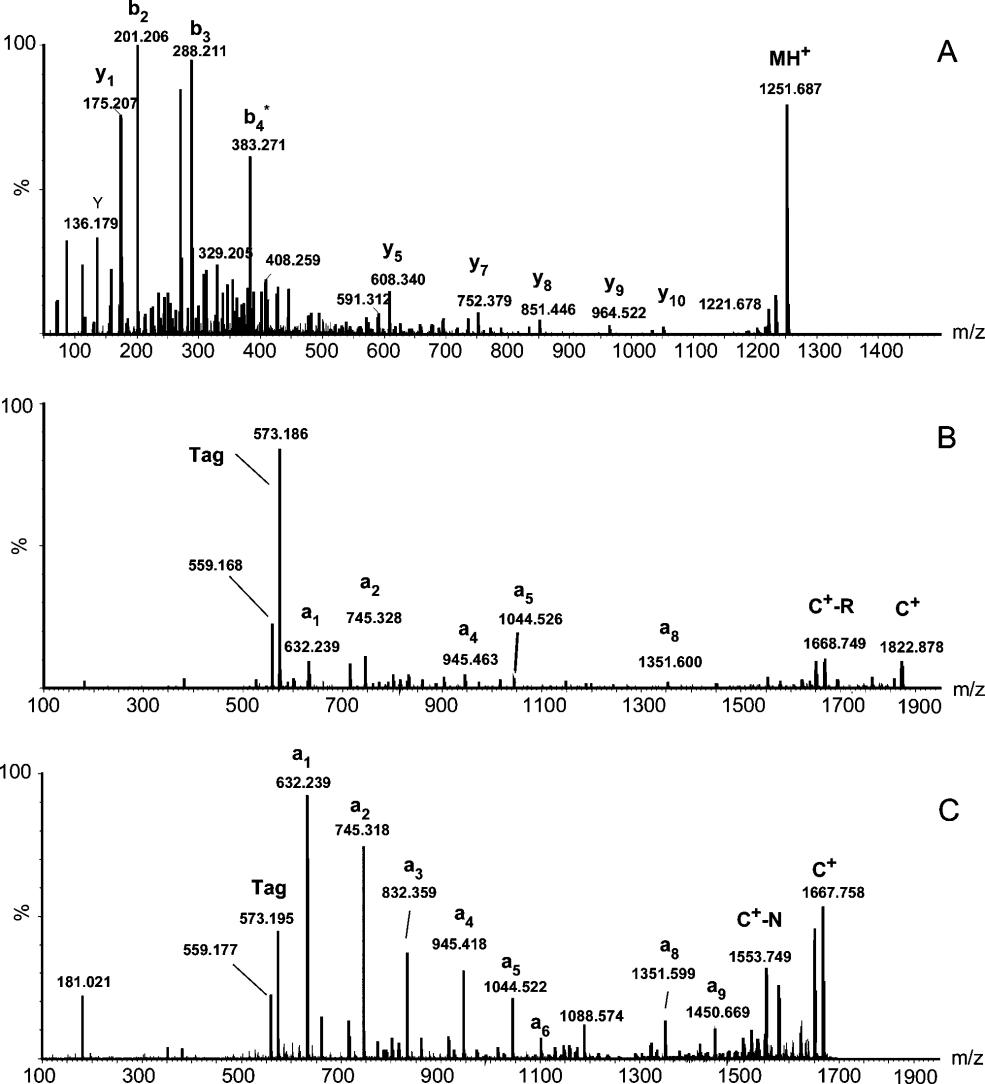 Figure 3. MALDI Q-TOF tandem mass spectra of a tryptic peptide, SISIVGSYVGNR, from alcohol degyhydragenases (ADH).
Figure 3. MALDI Q-TOF tandem mass spectra of a tryptic peptide, SISIVGSYVGNR, from alcohol degyhydragenases (ADH).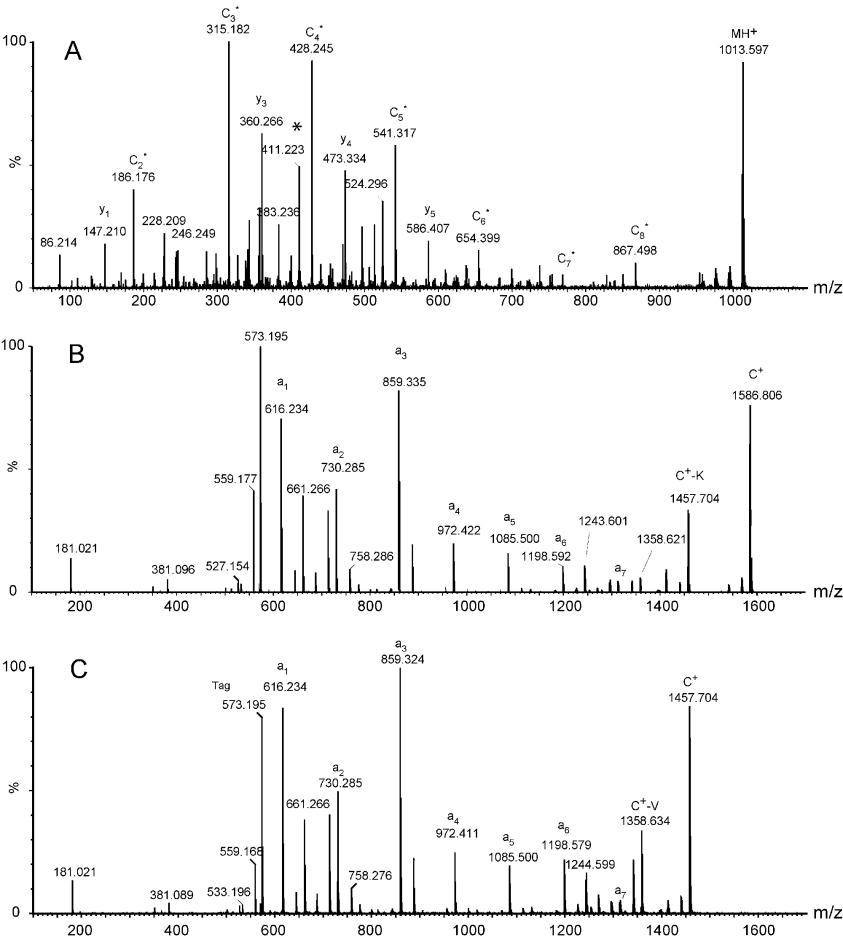 Figure 4. MALDI Q-TOF tandem mass spectra of a tryptic peptide, ANELLINVK, from alcohol degyhydragenases (ADH).
Figure 4. MALDI Q-TOF tandem mass spectra of a tryptic peptide, ANELLINVK, from alcohol degyhydragenases (ADH).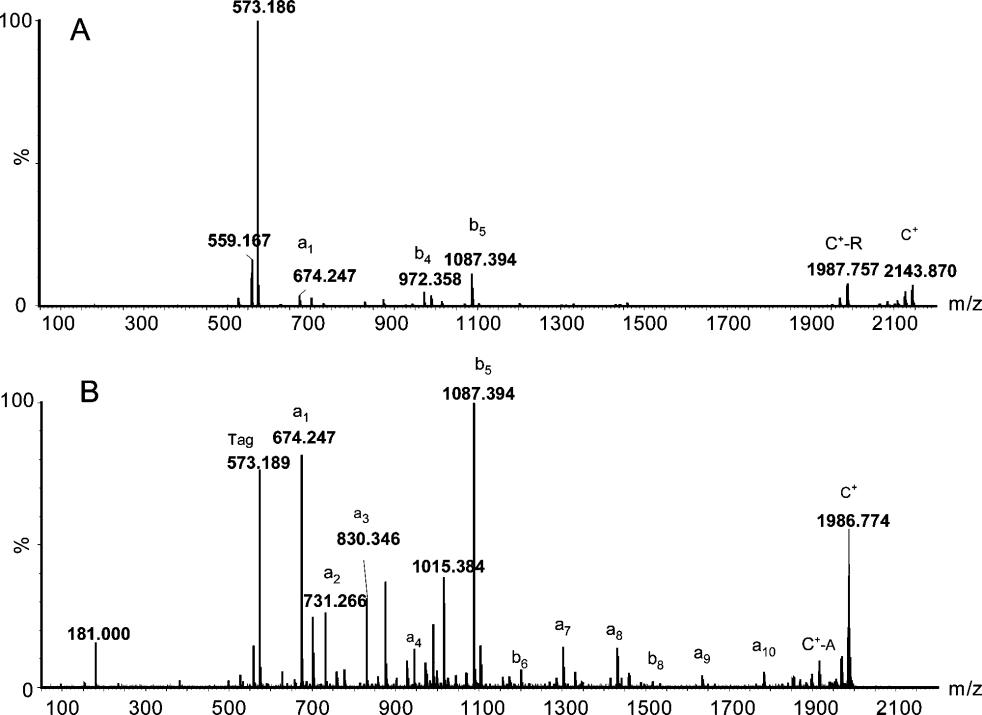 Figure 5. Comparison of MS/MS spectra of TMPP-Ac derivatives for a peptide with and without the presence of an Arg residue.
Figure 5. Comparison of MS/MS spectra of TMPP-Ac derivatives for a peptide with and without the presence of an Arg residue.References
- Chen W, et al. Improving de novo sequencing of peptides using a charged tag and C-terminal digestion. Analytical chemistry, 2007, 79(4): 1583-1590.
- Asakawa D. Principles of hydrogen radical mediated peptide/protein fragmentation during matrix-assisted laser desorption/ionization mass spectrometry. Mass Spectrom Rev, 2016.
- Zhang X, Zhang L, Li J. Peptide-modified nanochannel system for carboxypeptidase B activity detection. Anal Chim Acta, 2019.
- Bonetto V, et al. C-terminal sequence analysis of peptides and proteins using carboxypeptidases and mass spectrometry after derivatization of Lys and Cys residues. Analytical chemistry, 1997, 69(7): 1315-1319.
Related Services
Support Documents
KNOWLEDGE CENTER
KNOWLEDGE CENTER
KNOWLEDGE CENTER












