- Services
- FAQ
- Demo
- Case Study
- Related Services
- Support Documents
- Inquiry
What is Protein Sequence?
A protein sequence refers to the specific order in which amino acids are arranged in a protein chain. Each protein has a characteristic number and sequence of amino acid residues. The primary structure of a protein determines how the protein folds into a unique three-dimensional structure secondary, tertiary, and quaternary structures, which in turn determines the biological function of the protein. Protein sequences are critical for exploring mechanisms of diseases, designing therapeutic interventions, and advancing biotechnological applications.
The determination of protein sequence information can be used to design oligonucleotide probes complementary to the predicted gene sequence. These nucleic acid probes play an important role in cloning new genes encoding low-abundance proteins. For example, genes encoding many important drug proteins used in the clinic. Protein sequencing analysis can directly identify post-translational modifications that cannot be predicted from gene sequences, characterize recombinant proteins, and fragments of natural and recombinant proteins for structural and functional studies. Recombinant protein drugs can also be characterized to ensure that the expressed product conforms to the predicted structure, and to evaluate the quality and purity of the product to be highly consistent.
Creative Proteomics is a reliable partner of biopharmaceuticals. We provide reliable, high-quality protein sequence analysis service in accordance with relevant guidelines (ICH Q6B and USP<1047>) and perfect pre-sales and after-sales services.
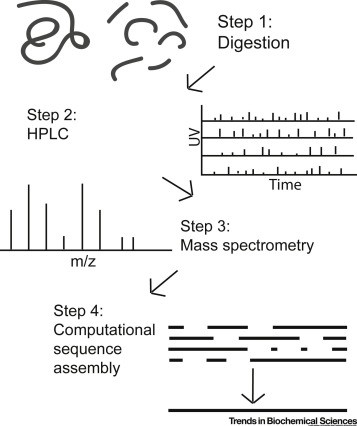 Figure 1. Current Protein Sequencing Paradigm. (Callahan N, et al. 2020)
Figure 1. Current Protein Sequencing Paradigm. (Callahan N, et al. 2020)Methods for Protein Sequencing
| Method | Principle | Advantages | Limitations |
|---|---|---|---|
| Mass Spectrometry | Identifies proteins by measuring the mass-to-charge ratio of ionized protein fragments. | - High sensitivity and accuracy - Can analyze complex mixtures - Suitable for PTM analysis |
- Requires specialized equipment - Sample prep can be complex |
| Edman Degradation | Sequentially removes and identifies N-terminal amino acids from peptides. | - High accuracy for N-terminal sequencing - Reliable for small proteins |
- Not suitable for large proteins - Requires pure protein samples |
| N-Terminal and C-Terminal Sequencing | Determines amino acid sequence at the N- or C-terminus of a protein using chemical or enzymatic cleavage. | - Provides terminal amino acid information - Useful for identifying protein fragments |
- Limited to terminal sequence data - May not provide complete protein sequence |
| Nanopore Protein Sequencing | Measures changes in ionic current as proteins pass through a nanopore, providing real-time sequencing. | - Real-time sequencing - High sensitivity - Minimal sample preparation |
- Still emerging technology - Requires specialized equipment |
We Can Provide (but not limited to):
- N-terminal Sequencing Service
- C-terminal Sequencing Service
- Peptide Mapping Service
- De novo Sequencing Service
- Monoclonal Antibody or Recombinant Protein Sequencing Service
- Amino Acids Analysis Service
- Amino Acid Composition Analysis Service
Technology Platform of Protein Sequencing
At Creative Proteomics, we utilize the latest and most advanced technology platforms for protein sequencing to ensure the highest accuracy, speed, and sensitivity in every project. Our technology suite includes:
High-Resolution Mass Spectrometry (LC-MS/MS): For deep proteomic analysis and detailed protein identification.
Edman Degradation Sequencing: For reliable and precise sequencing of small proteins and peptides.
Next-Generation Sequencing (NGS): For large-scale, high-throughput protein sequencing based on gene sequence information.
Nanopore Sequencing Technology: For real-time sequencing with minimal sample preparation.
 Figure 2. Mass spectrometry platform for protein sequencing.
Figure 2. Mass spectrometry platform for protein sequencing.Why Choose Our Protein Sequencing Service
- Rich Experience: Our company has been engaged in protein analysis of biopharmaceuticals for 16 years.
- Professional Platforms: High-resolution (greater than 105), high-quality accuracy (less than 1ppm) mass spectrometry platform and reverse phase high performance liquid chromatography (RP-HPLC) to ensure the accuracy of identification results.
- Bioinformatics Services: Our bioinformatics analysts can help you interpret the final data.
- Customized Service: You can choose the appropriate test item according to your project, and we will customize a unique solution for you.
Creative Proteomics' analytical scientists can provide quick turnover, clear and concise written reports and customized services to help customers solve analytical and technical problems.
Applications of Protein Sequencing
- Biomarker Discovery and Disease Research: Protein sequencing is instrumental in identifying novel biomarkers for disease detection and therapy monitoring.
- Therapeutic Protein Development: In the pharmaceutical industry, protein sequencing ensures the correct structure and functionality of therapeutic proteins. It is also critical in the development of biosimilars and in ensuring regulatory compliance for biologic drugs.
- Proteomics and Structural Biology: Protein sequencing plays a central role in structural proteomics, enabling the determination of 3D protein structures and interaction networks.
- Post-Translational Modification Analysis: Protein sequencing enables the detection and mapping of PTMs, which are essential for understanding protein function, stability, and interactions in cellular processes.
- Protein Engineering: By providing insights into protein sequences, protein sequencing aids in the engineering of novel proteins for various industrial applications, including enzyme design, antibody development, and therapeutic proteins.
Sample Requirements
| Sample Type | Required Amount | Purity | Storage |
|---|---|---|---|
| Protein Extract | 50 µg - 1 mg | > 90% pure | -20°C or -80°C (short-term) |
| Peptide Samples | 1 - 5 µg | > 90% pure | -20°C or -80°C (short-term) |
| Cell Lysate | 100 µg - 1 mg | High quality with minimal contaminants | -80°C (long-term storage) |
| Tissue Samples | 100 mg - 1 g | Cleaned and homogenized | Frozen (snap-freeze or -80°C) |
FAQ
Q: Can you perform protein sequencing on samples that have been previously frozen or stored for long periods?
A: Yes, we can perform protein sequencing on samples that have been frozen or stored for extended periods. However, it is important to ensure that the samples are stored properly (e.g., at -80°C for long-term storage) and have not experienced repeated freeze-thaw. We provide specific guidelines for sample preparation and storage to minimize degradation and ensure the integrity of the protein for sequencing.
Q: Do you offer any bioinformatics support for downstream analysis and interpretation of sequencing data?
A: Yes, we provide bioinformatics support to help with the downstream analysis of sequencing data. This includes sequence alignment, quantitative analysis, functional annotation, and interpretation of results in the context of biological pathways, disease mechanisms, or drug discovery. Our bioinformatics team can assist with identifying biomarkers, protein networks, and post-translational modifications.
Q: Do you offer any post-sequencing support, such as data analysis or interpretation services?
A: Yes, we offer comprehensive post-sequencing support, including:
Bioinformatics Analysis: Processing and analyzing sequencing data to extract meaningful insights.
Functional Annotation: Identifying and annotating proteins and their functions.
Visualization Tools: Providing tools to visualize data, such as protein interaction networks.
Consultation Services: Offering expert advice on experimental design and data interpretation.
Demo
Demo: Protein Identification by Tandem Mass Spectrometry and Sequence Database Searching.
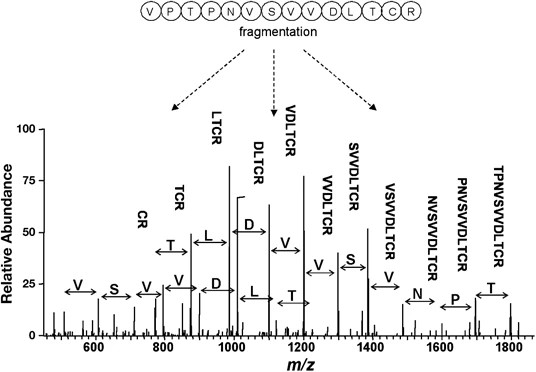 Figure 2. An example of a tandem mass spectrometry spectrum.
Figure 2. An example of a tandem mass spectrometry spectrum.Case Study
Case: Isolation and determination of the primary structure of a lectin protein from the serum of the American alligator (Alligator mississippiensis)
Background
Lectins are proteins that bind to carbohydrates with high specificity and are crucial for various biological functions, including immune defense and cell signaling. In mammals, lectins like intelectins play key roles in innate immunity and pathogen recognition. However, limited information is available on lectins from reptiles, particularly from the American alligator (Alligator mississippiensis). This study focuses on the isolation and characterization of a 35 kDa lectin from alligator serum, aiming to uncover its primary structure, carbohydrate-binding properties, and its role in immune defense.
Methods
- Protein Isolation: The lectin was isolated from alligator serum using affinity chromatography with mannose agarose to selectively bind mannose-binding proteins.
- Mass Spectrometry: The isolated lectin was analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS) and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) to determine the intact protein mass and to confirm the oligomeric forms of the lectin.
- Enzymatic Digestion: The lectin was enzymatically digested using various proteases (trypsin, Lys-C, Glu-C, Asp-N, and α-chymotrypsin) to generate peptides for LC-MS/MS analysis, enabling de novo sequencing of the protein.
- Edman Degradation: N-terminal sequencing of the protein was performed using Edman degradation to provide initial sequence data.
- Bioinformatics Analysis: The generated peptide sequences were analyzed using MASCOT Distiller and PEAKS software for de novo sequencing, and results were compared with the NCBI database using MASCOT and BLAST for protein identification.
Results
- Protein Characteristics: The 35 kDa lectin was found to exist primarily as a monomer and dimer in vitro, with oligomeric forms (trimer and tetramer) detected by mass spectrometry.
- Sequence Determination: Approximately 98% of the 35 kDa lectin sequence was successfully determined, revealing a total of 313 amino acids. The sequence showed 58%–59% similarity to intelectin-1 from humans and mice.
- Binding Affinity: The lectin exhibited strong binding to mannan and mannose, confirming its function as a mannose-binding lectin. Weaker binding was observed for other carbohydrates such as β-d-glucose, N-acetylglucosamine, and D-lactose.
- Structural Insights: The sequence analysis revealed a fibrinogen-related domain near the N-terminus, similar to the calcium-dependent carbohydrate recognition domain found in intelectins. However, the alligator lectin did not exhibit a consensus N-glycosylation site, unlike its human counterparts.
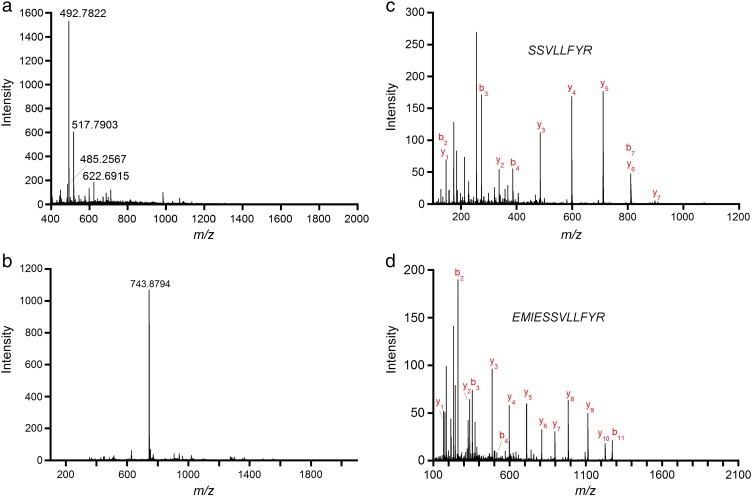 Figure 3. Mass spectrometry data for peptide sequencing.
Figure 3. Mass spectrometry data for peptide sequencing.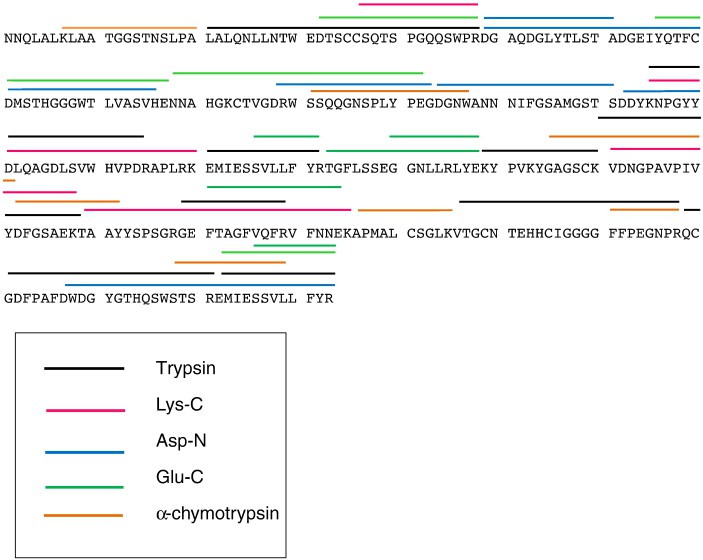 Figure 4. Primary structure of the 35 kDa lectin protein isolated from American alligator assembled from different endoprotease digestions.
Figure 4. Primary structure of the 35 kDa lectin protein isolated from American alligator assembled from different endoprotease digestions.References
- Callahan N, et al. Strategies for development of a next-generation protein sequencing platform. Trends in biochemical sciences, 2020, 45(1): 76-89. DOI: 10.1016/j.tibs.2019.09.005
- Nesvizhskii A I. Protein identification by tandem mass spectrometry and sequence database searching. Mass Spectrometry Data Analysis in Proteomics, 2007: 87-119. DOI: 10.1385/1-59745-275-0:87
- Darville L N F, et al. Isolation and determination of the primary structure of a lectin protein from the serum of the American alligator (Alligator mississippiensis). Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 2012, 161(2): 161-169. DOI: 10.1016/j.cbpb.2011.11.001
Related Services
Support Documents
KNOWLEDGE CENTER
KNOWLEDGE CENTER















