- Services
- FAQ
- Case Study
- Related Services
- Support Documents
- Inquiry
What is Near UV Circular Dichroism (CD) Spectroscopy
Circular dichroism (CD) spectroscopy is a fast, simple, and precise technique for analyzing protein structures in dilute solutions. This method relies on the principle that asymmetric molecules, such as proteins, absorb left and right circularly polarized light differently. By measuring the difference in absorption, CD spectroscopy provides insights into the secondary and tertiary structures of proteins, offering valuable data on protein folding, conformational changes, and stability.
Near UV CD Spectroscopy is a cutting-edge analytical technique used to explore the structural features and dynamic behaviors of proteins in solution. Operating within the 240 to 320 nm wavelength range, this method is highly sensitive to the electronic transitions of aromatic amino acids—such as tryptophan, tyrosine, and phenylalanine. By revealing subtle structural changes related to ligand binding, environmental shifts, or catalytic activity, near UV CD spectroscopy is essential in biochemistry and structural biology for understanding protein function and interactions.
All new biotechnology or biological products require spectral analysis as proof of conformity. As a reliable partner of biopharmaceutical companies, Creative Proteomics can provide customers with near UV circular dichroism spectroscopy analysis services based on GLP/GMP. The detectable molecules range from small chiral drugs and transition metal complexes to biological macromolecular structures such as proteins, glycoproteins and nucleic acids.
The Principles of Near UV CD Spectroscopy
Near UV CD Spectroscopy is grounded in the principle that chiral molecules, such as proteins, interact differently with left and right circularly polarized light. This technique exploits the unique electronic transitions of aromatic side chains in proteins—primarily tryptophan, tyrosine, and phenylalanine—by measuring the differential absorption of these polarized light beams.
Chirality of Proteins: Proteins are composed of amino acids, which can be chiral. The spatial arrangement of these chiral molecules leads to a preferential interaction with circularly polarized light. When a beam of polarized light passes through a solution containing proteins, the left and right circularly polarized light is absorbed to different extents based on the chirality of the protein.
Electronic Transitions: The aromatic amino acids absorb light due to π-π* electronic transitions. In near UV CD spectroscopy, the transitions of the aromatic side chains occur within the 240-320 nm wavelength range. The specific energies associated with these transitions are influenced by the local environment surrounding each aromatic residue, including interactions with other amino acids and the overall protein conformation.
Coupling of Excitations: The near UV CD signal is not solely dependent on the individual absorption of each aromatic residue but also on the coupling between the excitations of nearby aromatic groups. The proximity and relative orientation of these residues can enhance or diminish the observed CD signal, providing insights into their interactions.
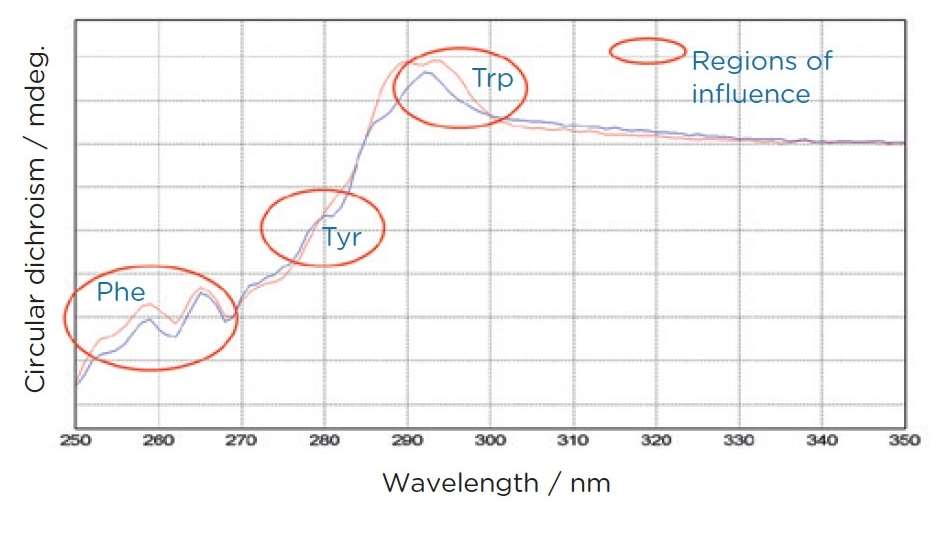
Spectral Features: The near UV CD spectrum comprises various peaks corresponding to the specific aromatic residues. Each residue contributes distinct signals based on its local conformation and interactions:
- Tryptophan typically exhibits signals around 284 and 297 nm.
- Tyrosine shows characteristic signals near 277 nm.
- Phenylalanine contributes signals around 261 and 268 nm. Additionally, disulfide bonds, while not aromatic, contribute broader signals that can influence the overall spectral profile.
We Can Provide but Not Limited to
At Creative Proteomics, we offer a wide range of near UV circular dichroism spectroscopy analysis services tailored to meet your needs:
- Environmental and Interaction Insights: Analyzing the interactions and environments of aromatic side chains to enhance your understanding of protein behavior.
- Binding Process Monitoring: Investigating ligand binding dynamics and the resulting conformational changes.
- Conformational Change Detection: Identifying structural variations from sequence modifications or environmental factors.
- Protein Folding Studies: Observing folding and unfolding processes under varying conditions to understand stability.
- Site-Directed Mutant Analysis: Evaluating contributions from individual aromatic residues in site-directed mutants to inform protein function.
Near UV CD Spectroscopy Analysis Workflow
At Creative Proteomics, we offer a wide range of near UV circular dichroism spectroscopy analysis services tailored to meet your needs:
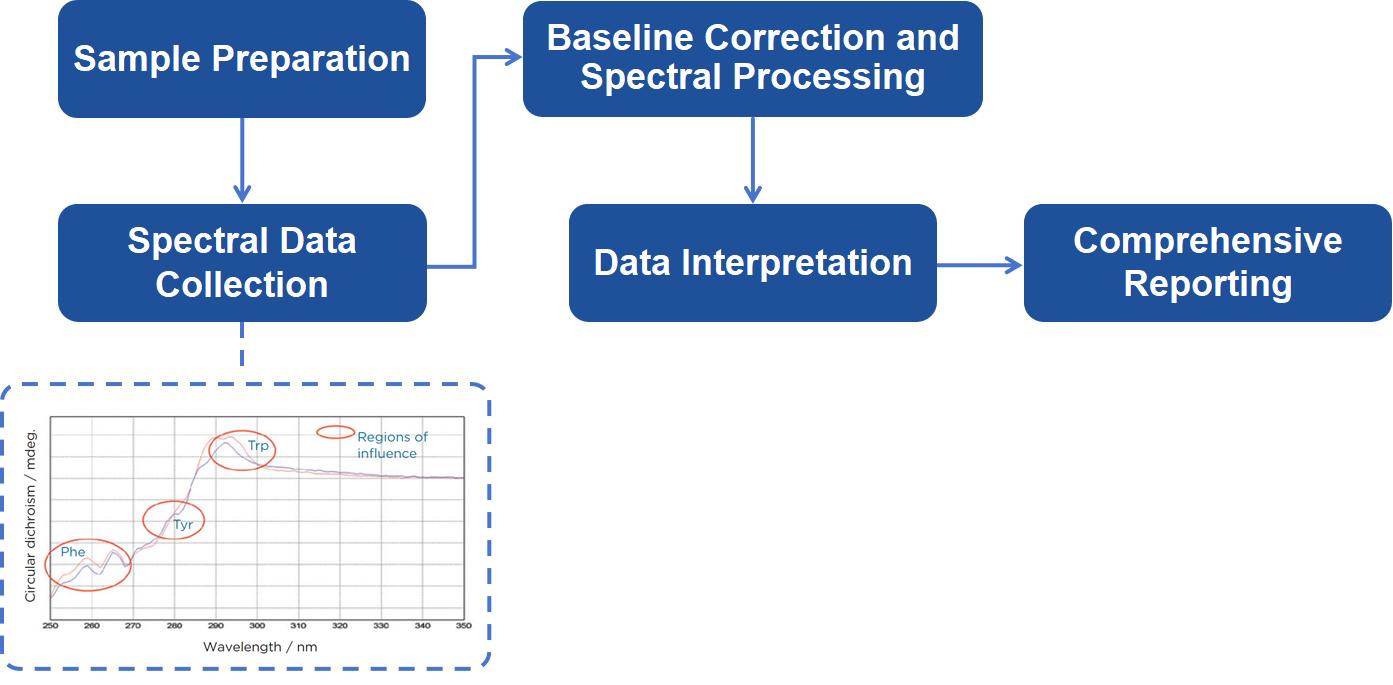
Sample Preparation: Preparing high-purity samples in appropriate buffers. Samples must meet specific criteria to ensure optimal data collection.
Spectral Data Collection: Using state-of-the-art CD spectrometers, we acquire spectra over the near UV region. Our equipment is calibrated to ensure the highest accuracy, and pathlengths are optimized for each sample concentration.
Baseline Correction and Spectral Processing: A baseline spectrum is acquired from the buffer alone, which is subtracted from the sample spectrum to yield a corrected CD signal. Further processing includes converting the signal into molar circular dichroism (Δε) to normalize for sample concentration and pathlength.
Data Interpretation: Our scientists interpret the CD spectra, focusing on the identification and quantification of secondary structural elements. We analyze results to extract structural information, focusing on aromatic interactions and conformational changes.
Comprehensive Reporting: After the analysis, we provide a detailed report that includes the CD spectra, a breakdown of secondary structure, and an interpretation of the data.
Sample Requirements
- Purity: The purity of samples for CD spectroscopy must be at least 90% (HPLC, MS or SDS-PAGE).
- Concentration: Sample concentration> 5 mg / ml.
- Buffer Conditions: Sample volume> 1 mL.
Creative Proteomics' analytical scientists can provide a clear and concise written report of the spectrum data of the far ultraviolet circular dichroism analysis to help customers analyze the secondary structure of the protein. We can also provide you with a one-stop service from sample preparation, purity analysis to circular dichroism analysis.
FAQ
Q: What kind of post-analysis support do you offer, particularly in terms of data interpretation?
A: After analysis, we provide a comprehensive report detailing the spectral data, alongside expert interpretations that contextualize the findings within your specific research framework. We encourage clients to schedule a follow-up consultation to discuss the results, where our scientists can answer questions and provide deeper insights into the implications of the data. This collaborative approach helps ensure that you fully understand how to leverage the results for your research.
Q: Can near UV CD spectroscopy be combined with other analytical techniques, and how would that enhance the analysis?
A: Yes, near UV CD spectroscopy can be effectively combined with other analytical techniques, such as fluorescence spectroscopy, mass spectrometry, or NMR spectroscopy. This multi-faceted approach allows for a more comprehensive understanding of protein structure and dynamics. For instance, while near UV CD provides insights into aromatic interactions and conformational states, fluorescence can offer information on overall protein folding and stability. By integrating data from multiple techniques, we can build a more complete picture of the protein's behavior and interactions.
Case Study
Case: Circular Dichroism Spectroscopy as a Tool for Monitoring Aggregation in Monoclonal Antibody Therapeutics
Background
Therapeutic Protein Aggregation: High-molecular-weight protein aggregates can affect the potency and stability of therapeutic proteins and may induce immune responses in patients.
Importance: Aggregates, especially those >100,000 Da, can alter the pharmacokinetics of therapeutic proteins and cause adverse reactions.
Study Objective: This study investigates multiple analytical methods for detecting and quantifying protein aggregates in recombinant acid alpha-glucosidase (rhGAA), a glycoprotein with an apparent molecular weight of 110 kDa.
Methods
- Protein Preparation: RhGAA was subjected to forced aggregation by incubation in sodium phosphate buffer at 37°C for 18 hours.
- Spiked Samples: A series of samples with varying aggregate concentrations (0%–20%) was created.
- Circular Dichroism (CD): Near-UV CD was used to detect changes in protein tertiary structure due to aggregation.
- Sedimentation Velocity Analytical Ultracentrifugation (AUC-SV): Samples were analyzed to quantify aggregation and assess the precision and limit of quantitation (LOQ).
- Size Exclusion Chromatography (SEC): Performed in parallel to analyze aggregate formation and compare results with AUC-SV.
- Purified Aggregates: Aggregates were purified from rhGAA finished products and compared to the forced aggregates.
Results
- rhGAA Aggregation: Incubation at neutral pH and elevated temperature led to irreversible formation of high-molecular-weight rhGAA aggregates.
- CD Sensitivity: Near-UV CD detected aggregation at levels as low as ≤4%, but far-UV CD did not show significant changes, indicating that the tertiary structure was more affected than the secondary structure.
- AUC-SV vs. SEC: AUC-SV provided better resolution of high-molecular-weight aggregates than SEC but had higher variability at low aggregate levels. SEC had a limit of quantitation (LOQ) of ≥0.2%, while AUC-SV had an LOQ of ≥3%.
- Precision: AUC-SV precision improved with higher aggregate levels (mean RSD 37.2%), while SEC showed higher overall precision (mean RSD 3.8%).
- Comparison of Methods: AUC-SV and SEC results were generally consistent, though AUC-SV showed a slight positive bias for aggregate content.
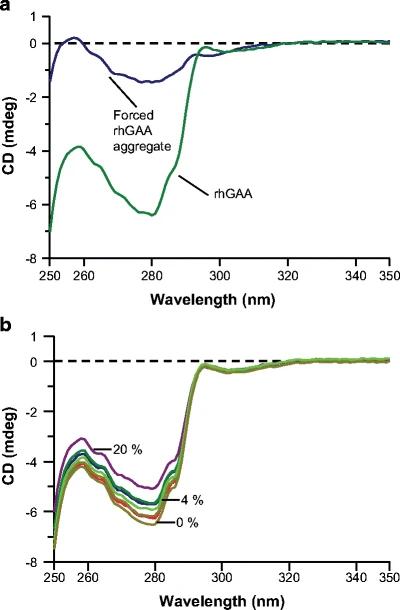 Figure 1. Near-UV circular dichroism spectra of (a) rhGAA forced aggregate versus rhGAA and (b) the rhGAA forced aggregate dilution series. Spectra for 0%, 4%, and 20% rhGAA forced aggregate levels are shown
Figure 1. Near-UV circular dichroism spectra of (a) rhGAA forced aggregate versus rhGAA and (b) the rhGAA forced aggregate dilution series. Spectra for 0%, 4%, and 20% rhGAA forced aggregate levels are shownReferences
- Hughes H, et al. A multi-tiered analytical approach for the analysis and quantitation of high-molecular-weight aggregates in a recombinant therapeutic glycoprotein. The AAPS journal, 2009, 11: 335-341.
- Li Z, Hirst J D. Quantitative first principles calculations of protein circular dichroism in the near-ultraviolet. Chemical science, 2017, 8(6): 4318-4333.
- Greenfield N J. Circular dichroism (CD) analyses of protein-protein interactions. Protein-Protein Interactions: Methods and Applications, 2015: 239-265.
Related Services
Support Documents
KNOWLEDGE CENTER
KNOWLEDGE CENTER












