- Services
- Demo
- Case Study
- FAQ
- Related Services
- Support Documents
- Inquiry
In biopharmaceutical development, understanding the integrity and structure of protein-based drugs is critical. From early R&D to quality control, each step demands accurate analytical methods. That's where MALDI-TOF-MS excels.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF-MS) enables fast, high-resolution analysis of intact proteins and peptides ranging from 1 to 100 kDa. This method delivers full-spectrum mass measurements without requiring deconvolution, offering significant advantages over traditional electrospray ionization (ESI) systems.
Within just five minutes, MALDI-TOF-MS can generate ion fragments that reveal structural characteristics—making it ideal for detecting covalent modifications in peptide-polymer conjugates, verifying drug stability, and profiling antibody formulations.
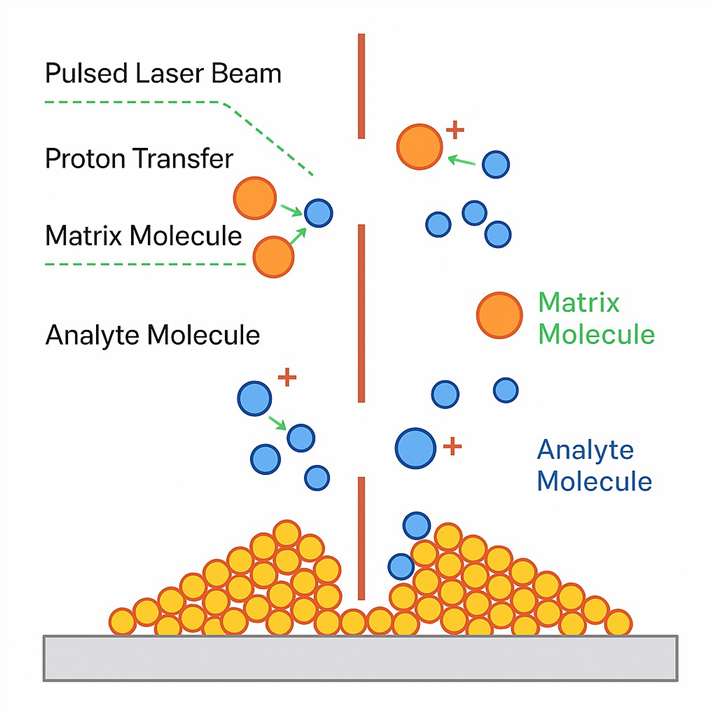 Matrix-assisted laser desorption/ionization.
Matrix-assisted laser desorption/ionization.Why Use MALDI-TOF-MS for Intact Protein Analysis?
Creative Proteomics offers comprehensive MALDI-TOF-MS services tailored for biologics developers. Our platform is designed for:
- Precise molecular weight determination
- Characterization of monoclonal antibodies (mAbs)
- Qualitative assessment of protein–polymer conjugates
- Monitoring physical and chemical stability in protein drugs
- Detecting covalent binding in biopharmaceutical formulations
Whether you're developing therapeutic peptides or engineering polymer-bound protein drugs, this method provides clear and rapid results.
Key Benefits of Our MALDI-TOF-MS Intact Protein Service
- High resolution (~20,000) and mass accuracy (<500 ppm)
- Exceptional sensitivity (~fmol to amol detection levels)
- High tolerance for detergents, buffers, and sample contaminants
- No need for complex deconvolution algorithms
- Fast turnaround with high throughput capability
This makes MALDI-TOF-MS particularly suited for laboratories working with polymeric protein complexes, or conducting lot-to-lot stability studies in preclinical and manufacturing settings.
Our Commitment:
Every MALDI-TOF-MS analysis is reviewed by a senior MS scientist to ensure accuracy and interpretability.
Note: MALDI-TOF is less effective for detecting low molecular weight species (<500 m/z). For such applications, ESI-MS may be more suitable.
| Feature | MALDI-TOF-MS | ESI-MS |
|---|---|---|
| Sample Tolerance | High (buffers, detergents) | Moderate |
| Analysis Speed | Fast (~5 min/sample) | Slower |
| Data Interpretation | Simple (less deconvolution) | Needs post-processing |
| Multi-Charge Ion Complexity | Low (cleaner spectrum) | High (complex patterns) |
| Low MW Detection (<500 m/z) | Not ideal | Excellent |
| Best Use Cases | Intact MW, conjugates, mAbs | PTM profiling, peptide mapping |
Workflow
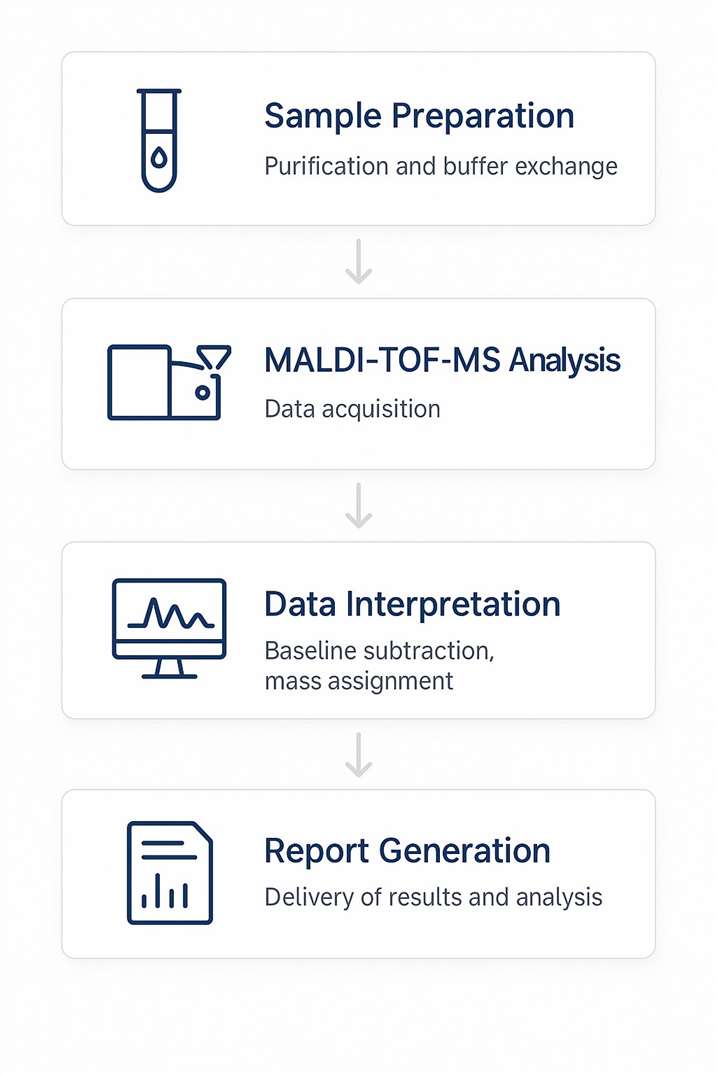
Sample Requirements
To ensure accurate and reproducible results, please prepare your samples following these guidelines:
| Parameter | Requirement |
|---|---|
| Purity | >90% |
| Mass Range | 1–100 kDa |
| Minimum Quantity | >40 µg |
| Preferred Buffers | Ammonium acetate, bicarbonate, or formate |
| Avoid | Tris, PBS, MES, HEPES, SDS, PEG, salts |
Avoid surfactants (e.g., Triton, CHAPS) and inorganic ions such as phosphate and sulfate, as they interfere with ionization.
Deliverables
With every MALDI-TOF-MS intact protein analysis, we provide:
Raw spectral data
- Clear, annotated results reports
- Optional bioinformatics interpretation
- Expert consultation for follow-up studies
Our platform is optimized for protein mass profiling in complex biopharmaceutical matrices
Demo
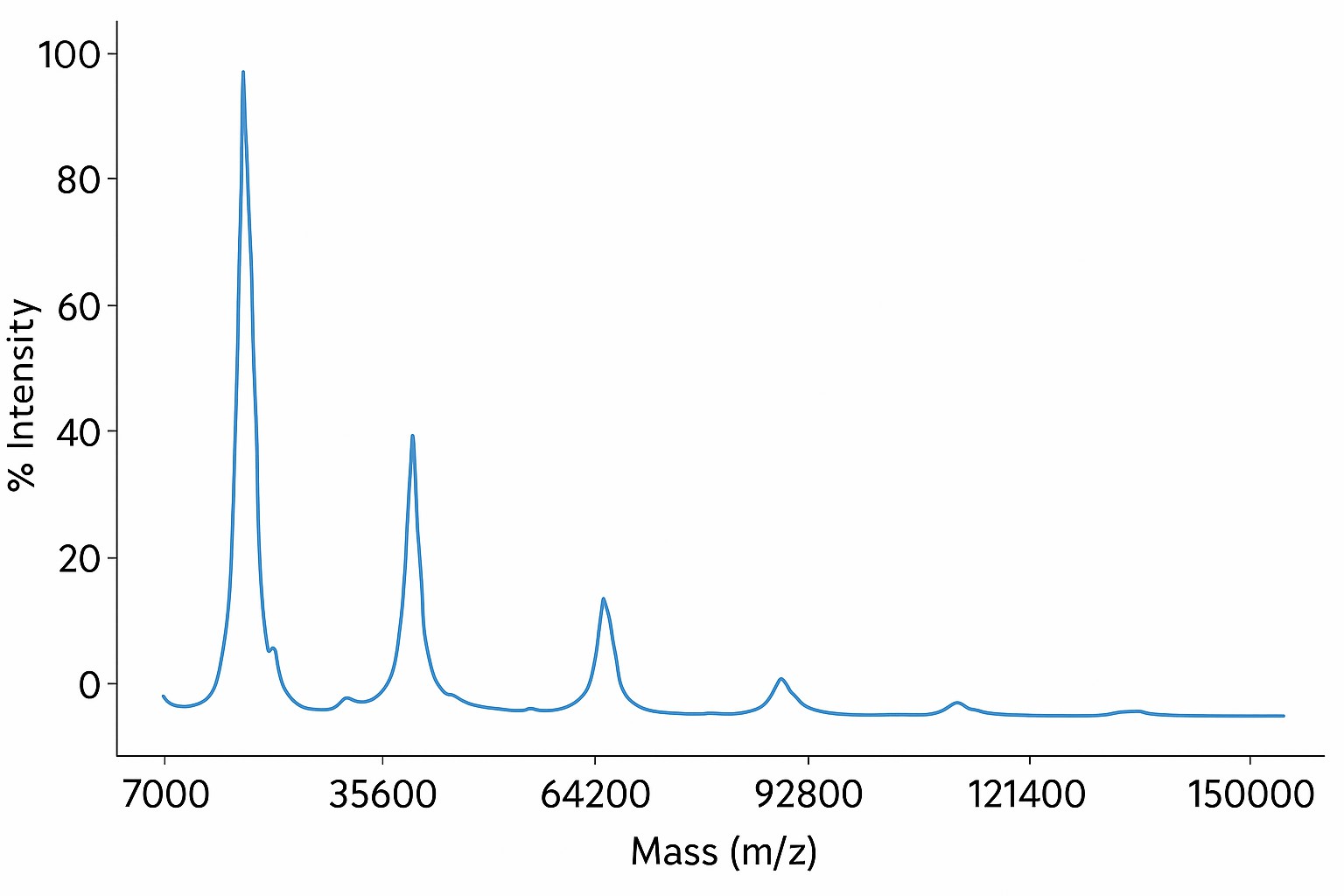
Case Study: Sequence-Independent Identification of SARS-CoV-2 Antibodies Using MALDI-TOF-MS
Tscheuschner, G., Kaiser, M.N., Lisec, J. et al. MALDI-TOF-MS-Based Identification of Monoclonal Murine Anti-SARS-CoV-2 Antibodies within One Hour. Antibodies 2022, 11(2), 27.
DOI: 10.3390/antib11020027
Background
The reproducibility crisis in antibody-based research has highlighted the need for fast and reliable methods to confirm antibody identity. Traditional sequencing approaches are costly and often infeasible. In this context, a research team from Germany (BAM, RKI, Bruker, HybroTec) demonstrated that MALDI-TOF-MS can be used to identify monoclonal antibodies in under one hour, without the need for sequence data.
They tested this on 35 murine anti-SARS-CoV-2 monoclonal antibodies produced against various spike protein and nucleocapsid antigens.
Method Overview
Researchers used a multi-layered MALDI-TOF-MS workflow that included:
- Intact Mass Measurement
Analysis of the whole antibody to distinguish based on overall mass. - Light Chain Mass Profiling
Reduction of disulfide bonds followed by MALDI-TOF detection of the light chain (~23 kDa). - Peptide Mass Fingerprinting (PMF)
Rapid digestion using: - Diluted sulfuric acid (30 min, 99°C, no alkylation)
- Fast trypsin digestion (15 min at 55°C, no chaotropes)
- Combined peptide fingerprints gave >80% sequence coverage for the NIST-mAb reference.
Key Results
- 21 of 36 antibodies were uniquely identifiable by intact mass and light chain mass alone.
- Fingerprints from sulfuric acid and trypsin digestion were highly distinctive, enabling:
- Discrimination of antibodies from different immunizations
- Confirmation of identity between sister clones (e.g., 1008 & 1043)
- ABID 2.0, an open-source software, enabled automatic spectral matching against a virtual fingerprint library.
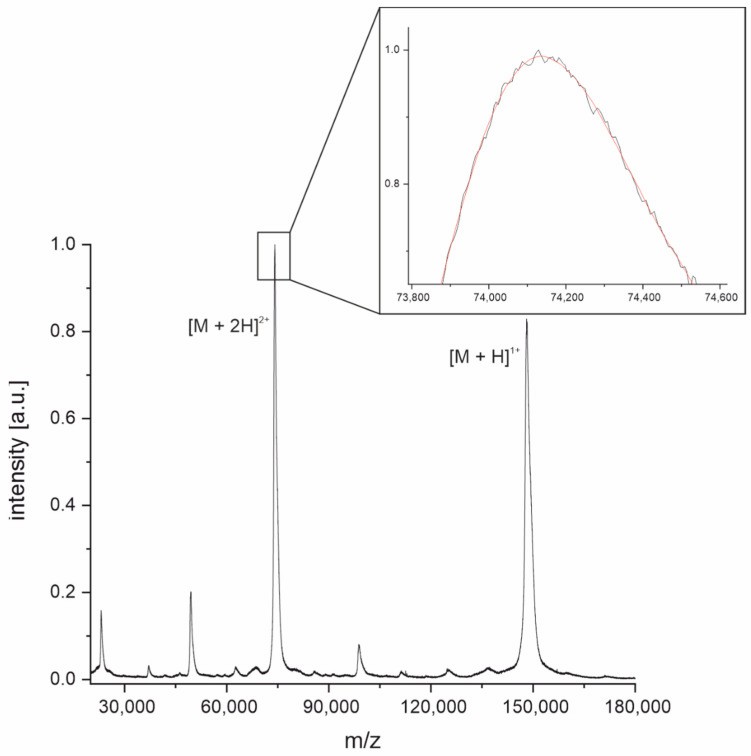 MALDI-TOF mass spectrum of intact NIST-mAb 8671.
MALDI-TOF mass spectrum of intact NIST-mAb 8671.Why This Matters for Protein Mass Services
This case shows that MALDI-TOF-MS isn't just for molecular weight analysis—it can also provide sequence-specific fingerprints, even without traditional sequencing. For clients developing monoclonal antibodies or biosimilars, this means:
- Rapid identity verification
- Reduced risk of clone mislabeling
- Sequence-independent QC documentation
Creative Proteomics can replicate and adapt such workflows for client-specific applications using MALDI-TOF-MS intact protein analysis, especially in mAb characterization, clone screening, and batch-to-batch identity checks.
Frequently Asked Questions (FAQs) – MALDI-TOF-MS Intact Protein Analysis Service
Q: What types of samples are suitable for MALDI-TOF-MS intact protein analysis?
A: This method is ideal for proteins and peptides ranging from 1 kDa to 100 kDa, including monoclonal antibodies, recombinant proteins, and polymer–protein conjugates. The technique works best with purified samples (>90%) in MS-compatible buffers.
Q: What level of sample purity is required?
A: For optimal results, samples should have a purity of at least 90%. Lower purity samples may produce interfering peaks or inaccurate results. If needed, we can discuss pre-cleanup or buffer exchange strategies.
Q: How much sample do I need to submit?
A: We recommend submitting at least 40 µg of protein per sample. If you're working with limited quantities, please contact us for consultation—our team can often adjust protocols for smaller inputs.
Q: What buffer systems are compatible with MALDI-TOF-MS?
A: Compatible buffers include:
- Ammonium acetate
- Ammonium bicarbonate
- Ammonium formate
Avoid using:
- Tris, PBS, MES, HEPES
- Surfactants like SDS, Tween, CHAPS
- Salts such as phosphates, sulfates, or halides
Q: Can you analyze complex samples like protein–polymer conjugates or modified antibodies?
A: Yes. MALDI-TOF-MS is especially suited for:
- Covalent protein–polymer conjugates
- PEGylated peptides
- Monoclonal antibodies (mAbs) We provide full molecular weight profiling to confirm modification success and product integrity.
Q: How does MALDI-TOF-MS compare with ESI-MS?
| Feature | MALDI-TOF-MS | ESI-MS |
|---|---|---|
| Speed | Fast | Moderate |
| Data Complexity | Simple (no deconvolution needed) | Complex (multi-charged ions) |
| Sample Tolerance | High | Medium |
| PTM Detection | Moderate | High |
| Best Use Case | Intact MW of stable proteins | Peptides, PTMs, low MW detection |
We may recommend ESI-MS if you're targeting low-mass peptides or PTMs. For intact proteins and polymer-bound drugs, MALDI-TOF is preferred.
Q: What kind of data will I receive in the final report?
A: Each report includes:
- Raw spectra files
- Peak table with molecular weights
- Spectral annotation (m/z, signal intensity)
- Interpretation of detected molecular forms
- Optional bioinformatics insights (e.g., glycoforms, dimerization, conjugates)
Q: How do I submit samples for MALDI-TOF-MS analysis?
A: Follow these steps:
- Contact our technical team to confirm buffer compatibility
- Fill out a sample submission form
- Package samples using ice packs or lyophilization
- Ship to our NY-based lab with tracking information
References
- Kafka A P, Kleffmann T, et al. The application of MALDI TOF MS in biopharmaceutical research. International journal of pharmaceutics, 2011, 417(1-2): 70-82. DOI: 10.1016/j.ijpharm.2010.12.010
- Staub A, Guillarme D, et al. Intact protein analysis in the biopharmaceutical field. Journal of pharmaceutical and biomedical analysis, 2011, 55(4): 810-822.DOI: 10.1016/j.jpba.2011.01.031
- Shubhakar A, Kozak R P, et al. Automated high-throughput permethylation for glycosylation analysis of biologics using MALDI-TOF-MS. Analytical chemistry, 2016, 88(17): 8562-8569. https://doi.org/10.1021/acs.analchem.6b01639
Related Services
Support Documents
KNOWLEDGE CENTER
Molecular Weight Characterization - Comparison of MALDI and ESI











