- Services
- FAQ
- Case Study
- Related Services
- Support Documents
- Inquiry
What is N-terminal Sequencing?
N-terminal sequencing is a pivotal analytical method employed to determine the amino acid sequence at the N-terminus of a protein. The N-terminus, characterized by the free amino group at the protein's outset, plays a crucial role in governing the protein's biological function, stability, and subcellular localization. By elucidating the N-terminal sequence, it offers critical insights into the protein's structural features, post-translational modifications, and functional properties.
N-terminal sequence analysis of proteins and peptide drugs is crucial for pharmaceutical quality control, aligning with ICH Q6B guidelines requiring N-segment sequence data. At Creative Proteomics, we provide comprehensive, GLP/cGMP-compliant N-terminal sequencing services, adhering to ICH standards and the US FDA's "Points to Consider" document, ensuring reliable and regulatory-compliant results.
Technology Platform of N-terminal Sequencing Analysis Service
Creative Proteomics effectively identifies and analyzes the N-terminus of proteins by fluorescent labeling, trypsin digestion, chemical labeling combined with mass spectrometry, and differential peptide mapping, etc.
We provide the following analysis methods:
1) N-terminal sequencing via Edman degradation
The N-terminal amino acid group of the protein is labeled with phenyl isothiocyanate (PITC), selectively cleaves the labeled N-terminal amino acid, and the extracted phenylthiohydantoin is identified by MS Urea (PTH)-amino acid, according to this procedure to determine the amino acid sequence of the protein. The specific workflow is as follows.
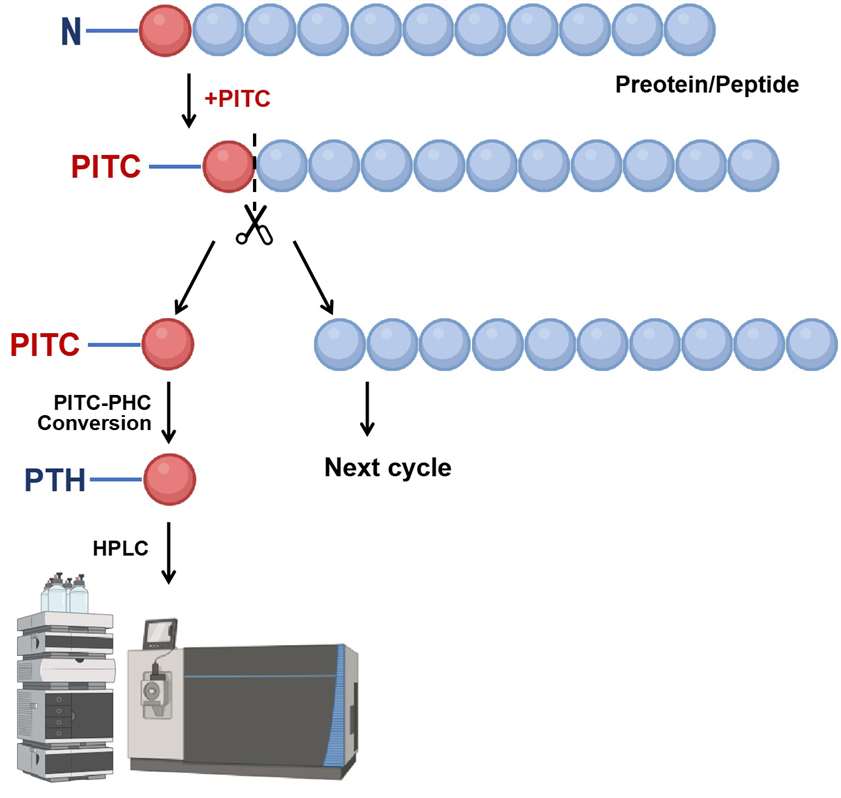 Figure 1. The workflow of the N-terminal sequencing via Edman degradation.
Figure 1. The workflow of the N-terminal sequencing via Edman degradation.2) De novo N-terminal peptide sequencing by Tandem MS
This technology employs fluorescent reagents to label the N-terminal α-amino group of proteins. After trypsin digestion, LC-MS/MS and fluorescence detection perform tandem mass spectrometry analysis. A differential peptide mapping step identifies pyroglutamate-blocked N-terminal peptides for high-quality sequencing. The specific workflow is as follows.
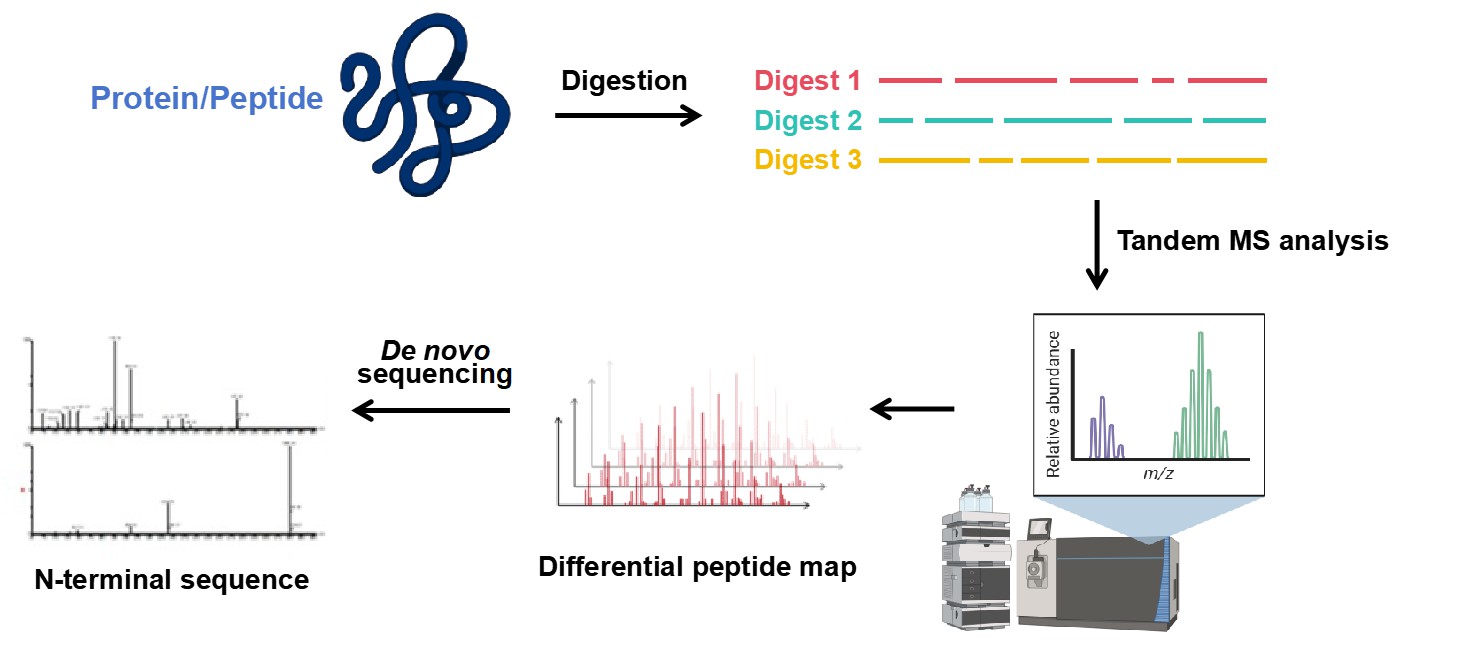 Figure 2. Workflow for De novo N-terminal sequencing by Tandem MS.
Figure 2. Workflow for De novo N-terminal sequencing by Tandem MS.Difference Between N-terminal Sequencing and C-terminal Sequencing
| Feature | N-terminal Sequencing | C-terminal Sequencing |
|---|---|---|
| Primary Technique | Edman degradation and tandem mass spectrometry (LC-MS/MS). | Carboxypeptidase digestion and mass spectrometric mapping. |
| Handling of Blocked Termini | Employs differential peptide mapping to resolve blocked or modified N-termini (e.g., pyroglutamate). | Can analyze ragged ends but lacks direct sequencing methods for blocked C-termini. |
| In Monoclonal Antibody Analysis | Identifies the N-terminal amino acids of light and heavy chains. | Evaluates variability due to removal of C-terminal lysine residues in heavy chains. |
| Advantages | Highly precise, resolves isomeric residues (e.g., leucine vs. isoleucine), supports detailed PTM analysis. | Useful for assessing structural variability and heterogeneity of C-terminal regions. |
| Applications | Protein identification, verification of batch-to-batch consistency, resolution of leucine/isoleucine. | Analysis of intactness, structural heterogeneity, and variations such as C-terminal lysine removal. |
Applications of N-terminal Sequencing
- Protein Identification: Verifies N-terminal sequences of biopharmaceuticals to meet ICH Q6B guidelines.
- Batch Consistency: Ensures reproducibility in biologics manufacturing.
- Post-Translational Modifications: Detects modifications like cyclization, methylation, and pyroglutamate formation affecting stability and efficacy.
- Regulatory Compliance: Provides required sequence data for therapeutic protein quality assurance.
- Isomeric Residue Resolution: Differentiates leucine and isoleucine, surpassing mass spectrometry's capabilities.
We Can Provide but Not Limited to:
- Identification of advanced structure of proteins
- N-terminal sequence analysis
- Analysis of N-terminal modification sites
- Sequencing light and heavy chain terminals
Advantages of Our N-terminal Sequencing Analysis Service
- Time Efficiency: Utilizing advanced platforms with simple sample preparation, the technology significantly reduces identification and analysis time.
- High Sensitivity: Combines fluorescent labeling and mass spectrometry for sensitive detection of N-glycosylation sites.
- High Throughput: Edman sequencing ensures precise N-terminal analysis, while mass spectrometry addresses N-terminal blockage and PTM challenges. For this reason, there is no limitation of sample types for protein N-terminal sequencing, and Will not be restricted by N-terminal block and PTM.
- Customized service: We can customize professional solutions for you according to your research plan needs. You can select or suggest the required items for analysis.
Creative Proteomics's professional researchers can provide customers with comprehensive N-terminal sequence sequencing information of biopharmaceutical proteins to realize the analysis of protein advanced structure and modification sites. We will provide you with detailed experimental process steps and the final N-terminal sequence information analysis report and other data reports. We are committed to providing you with expertise in protein N-terminal sequences to help you solve problems related to analytical techniques.
FAQ
Q: How does N-terminal sequencing handle blocked or modified termini?
A: Blocked or modified N-termini, such as pyroglutamate or acetylation, cannot be analyzed directly using Edman degradation. To overcome this, we employ complementary methods like:
De novo sequencing via tandem mass spectrometry (LC-MS/MS): This technique identifies blocked N-termini using differential peptide mapping.
Enzymatic or chemical deblocking strategies: These can remove specific blocking groups to enable sequencing.
Q: How does N-terminal sequencing contribute to PTMs analysis?
A: N-terminal sequencing identifies and characterizes PTMs at or near the N-terminus, including:
Glycosylation, phosphorylation, and acetylation.
Removal of signal peptides or leader sequences.
Variations arising from proteolytic processing.
Q: Can N-terminal sequencing analyze mixtures of proteins?
A: Yes, mixtures of proteins can be analyzed if they are first separated into individual components. Common separation methods include:
SDS-PAGE followed by transfer to a PVDF membrane for isolation of individual protein bands.
Chromatographic techniques such as HPLC.
Once separated, each protein can be sequenced individually using Edman degradation or LC-MS/MS.
Q: What is the sensitivity of N-terminal sequencing?
A: N-terminal sequencing is highly sensitive:
Edman degradation can detect picomolar to low nanomolar quantities of protein or peptide.
Tandem MS offers even higher sensitivity and can analyze samples with extremely low concentrations.
The sensitivity depends on the sample purity, quality, and the specific sequencing method employed.
Case Study
Case: N-terminal sequence analysis of equine herpesvirus 1 glycoproteins D and B and evidence for internal cleavage of the gene 71 product
Background
This study examines the cleavage sites of equine herpesvirus 1 (EHV-1) glycoproteins D and B (gD and gB) and the gene 71 product, crucial for viral infection, pathogenesis, and immune response. These glycoproteins undergo signal peptide and endoproteolytic cleavage for maturation and function. While conserved cleavage motifs were suggested in gB homologs of other herpesviruses, EHV-1 cleavage sites were only predicted, not experimentally confirmed. The research aims to experimentally identify the cleavage sites of EHV-1 gD and gB and evaluate the role of glycosylation in their processing.
Methods
Protein Purification: Glycoproteins were extracted from infected cell lysates and purified using immunoaffinity chromatography with monoclonal antibodies specific to gD, gB, and gene 71 products.
Western Blotting: Proteins were separated via SDS-PAGE, transferred to membranes, and identified using monoclonal antibodies.
N-terminal Sequencing: Purified proteins were transferred to a PVDF membrane, excised, and analyzed via Edman degradation for sequence determination.
Glycosylation Inhibition: Tunicamycin and monensin inhibited glycosylation, and its effects on gB cleavage were analyzed using Western blotting.
Results
The signal cleavage site for EHV-1 gD was experimentally determined between Arg35 and Ala36, nine residues downstream from the predicted site. For EHV-1 gB, cleavage occurred as predicted between Ala85 and Val86.
Internal cleavage of gB occurred between Arg548 and Ala549, 28 residues downstream of the predicted site. This cleavage site is unique to EHV-1 and does not align with the conserved cleavage motif found in other herpesviruses.
Cleavage of gB was unaffected by glycosylation inhibitors (tunicamycin and monensin), indicating that proteolytic cleavage can occur independently of glycosylation.
A 42 kDa protein derived from the product of gene 71 was identified as a cleavage product. The cleavage occurred between adjacent arginine residues (Arg506 and Arg507).
Immunoblotting confirmed the identities of glycoproteins and their subunits. The large and small subunits of gB were approximately 76 kDa and 54 kDa, respectively, and were linked by disulfide bonds in the native heterodimer.
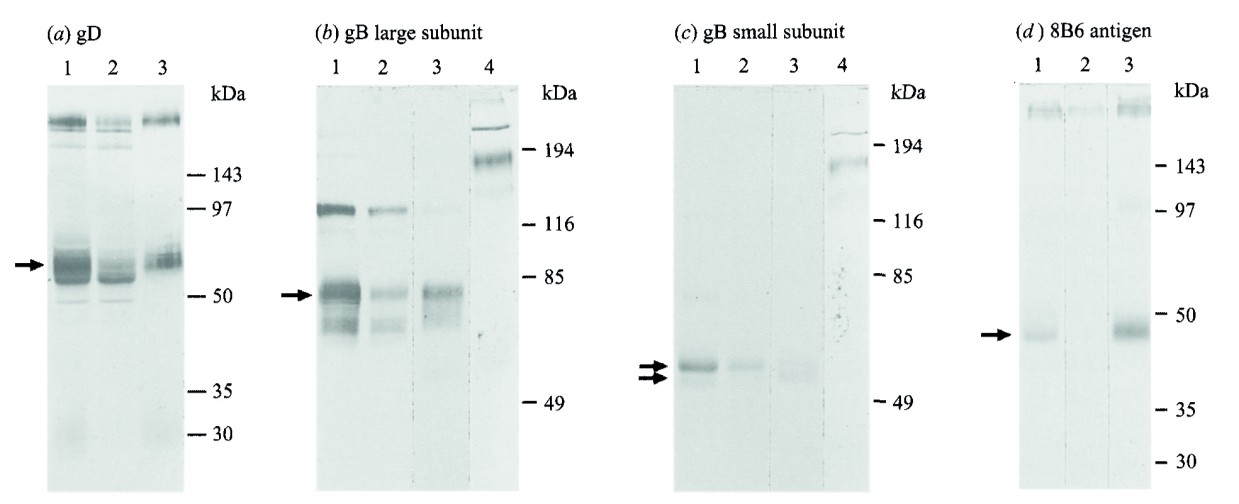 Figure 3. Identification by electrophoresis and immunoblotting of EHV-1 glycoproteins purified with MAbs 1F12 (anti-gD), 7B10 (anti-gB large subunit) and 8B6. Bands used for N-terminal sequencing.
Figure 3. Identification by electrophoresis and immunoblotting of EHV-1 glycoproteins purified with MAbs 1F12 (anti-gD), 7B10 (anti-gB large subunit) and 8B6. Bands used for N-terminal sequencing.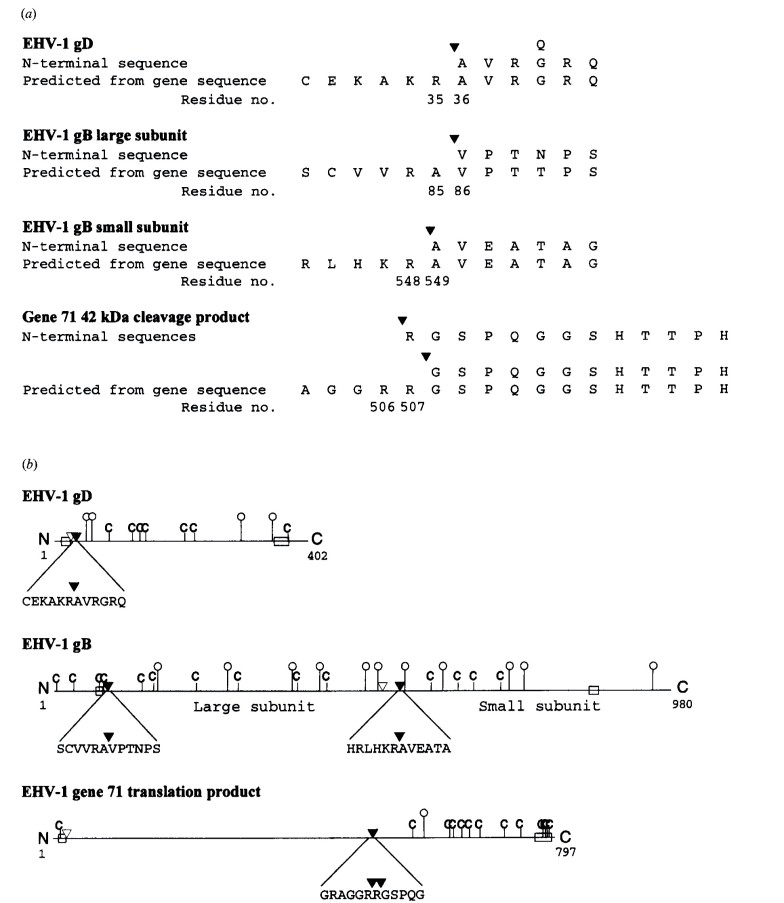 Figure 4. (a) Amino acid sequences of N termini of processed EHV-1 gD, small and large subunits of EHV-1 gB, and the 42 kDa cleavage product of gene 71, aligned with amino acid sequences predicted from nucleotide sequences. (b) Schematic diagram representing translation products of the EHV-1 genes for gD and gB and EHV-1 gene 71.
Figure 4. (a) Amino acid sequences of N termini of processed EHV-1 gD, small and large subunits of EHV-1 gB, and the 42 kDa cleavage product of gene 71, aligned with amino acid sequences predicted from nucleotide sequences. (b) Schematic diagram representing translation products of the EHV-1 genes for gD and gB and EHV-1 gene 71.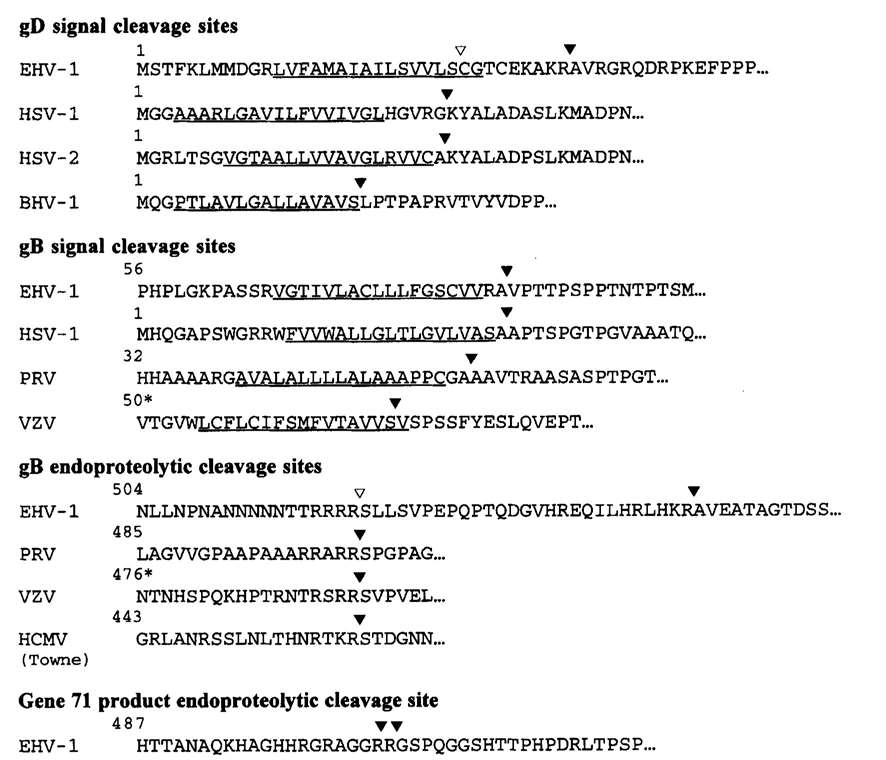 Figure 5. Comparison of signal and endoproteolytic cleavage sites of EHV-1 glycoproteins with homologous glycoproteins of other herpesviruses.
Figure 5. Comparison of signal and endoproteolytic cleavage sites of EHV-1 glycoproteins with homologous glycoproteins of other herpesviruses.References
- Yun S H, Choi C W, Lee S Y, et al. A proteomics approach for the identification of novel proteins in extremophiles. Biotechnology of Extremophiles: Advances and Challenges, 2016: 303-319. DOI: 10.1007/978-3-319-13521-2_10
- Vecchi M M, Xiao Y, Wen D. Identification and sequencing of N-terminal peptides in proteins by LC-fluorescence-MS/MS: An approach to replacement of the Edman degradation. Analytical chemistry, 2019, 91(21): 13591-13600. DOI: 10.1021/acs.analchem.9b02754
- Misal S A, Li S, Tang H, et al. Identification of N‐terminal protein processing sites by chemical labeling mass spectrometry. Rapid Communications in Mass Spectrometry, 2019, 33(11): 1015-1023. DOI: 10.1002/rcm.8435
- Wellington J E, Gooley A A, Love D N, et al. N-terminal sequence analysis of equine herpesvirus 1 glycoproteins D and B and evidence for internal cleavage of the gene 71 product. Journal of General Virology, 1996, 77(1): 75-82. DOI: 10.1099/0022-1317-77-1-75
Related Services
Support Documents












