The structure and function of proteins are intricately interlinked, with disulfide bonds serving as crucial chemical linkages between cysteine residues. These bonds play a pivotal role in maintaining protein stability, conformation, and biological activity. In antibody-based therapeutics, the precise formation and distribution of disulfide bonds are critical determinants of structural integrity and therapeutic efficacy. However, aberrant reduction or mispairing of these bonds can lead to protein inactivation, aggregation, and even immunogenicity risks, posing significant challenges to biopharmaceutical manufacturing processes. This article comprehensively examines the biological characteristics of disulfide bonds, their functional mechanisms in antibodies, and provides a full-spectrum strategy from formation regulation to reduction prevention. These insights serve as a scientific foundation for quality control in the development of antibody therapeutics.
What Is Disulfide Bonds
In chemistry, a disulfide bond represents a functional group characterized by the linkage R-S-S-R', typically formed by the covalent coupling of two thiol groups. In biological systems, disulfide bonds create covalent linkages either between separate peptide chains or between two cysteine thiol groups within a single peptide chain. These bonds are formed through the oxidative dehydration of free thiol groups from cysteine residues, resulting in the amino acid cystine. Disulfide bonds are pervasive in proteins, ranging from those in simple unicellular prokaryotes to those in complex multicellular eukaryotes. They play an essential role in stabilizing protein structure by maintaining conformational integrity.
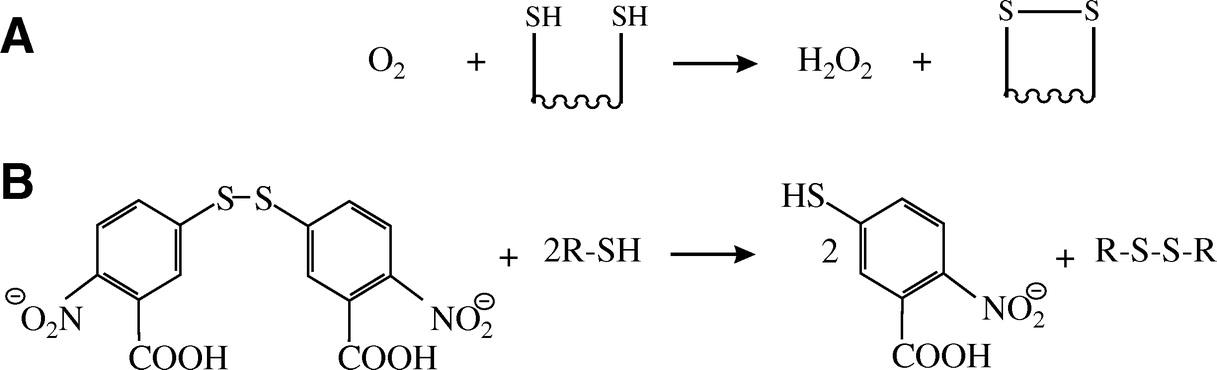 Schematic representation of the formation of a disulfide bond
Schematic representation of the formation of a disulfide bondProperties of Disulfide Bonds
Disulfide bonds, categorized as covalent interactions, significantly contribute to the stability of protein molecules by playing a pivotal role in maintaining their structural integrity. These bonds are known to enhance a protein's robustness when subjected to external perturbations, particularly when compared to other chemical bonds. The stability of a protein's conformation is directly proportional to the number of disulfide bonds it contains. Despite their stabilizing role, disulfide bonds exhibit dynamic flexibility rather than being statically rigid. These bonds are susceptible to cleavage under particular conditions, such as the presence of reducing agents. This inherent flexibility is crucial for modulating protein function, facilitating structural and functional adaptability of proteins in response to variations in the intracellular environment, thereby allowing them to meet diverse physiological requirements.
Function of Disulfide Bonds in Protein Structure
In the realm of protein structural biology, disulfide bonds are regarded as specialized covalent linkages occurring between specific amino acids, predominantly cysteine residues. These bonds are critically involved in preserving the integrity of the primary protein structure. At more advanced structural levels, disulfide bonds are indispensable for the stabilization of the spatial configuration of peptide chains, thereby serving as essential constituents in the formation of both secondary and tertiary structures. The three-dimensional conformation of proteins is a cumulative result of non-covalent interactions, including electrostatic forces, van der Waals forces, hydrogen bonds, and hydrophobic interactions, alongside covalent bonds such as disulfide linkages. Thus, disulfide bonds exhibit a profound connection to the amino acid sequence of the primary structure while remaining vital for the assembly and stabilization of the protein's higher-order architecture.
Disulfide Bond Structure and Function in Antibodies
Disulfide Bond Linkages in IgG Subclasses
Monoclonal immunoglobulin G (IgG) antibodies, predominantly expressed in Chinese hamster ovary (CHO) cells, consist of four subclasses: IgG1, IgG2, IgG3, and IgG4. The architecture of disulfide bond linkages within these subclasses was initially elucidated in the 1960s. Below is a summary table detailing the comparison of disulfide bond linkages across the four IgG subclasses:
| IgG Subclass | Intrachain Disulfide Bonds | Interchain Disulfide Bonds | Interchain Disulfide Bond Locations |
| IgG1 | 12 pairs | 2 pairs (linked through the hinge region) | Heavy chain: hinge region; Light chain and heavy chain: Light chain's terminal cysteine and heavy chain's fifth cysteine |
| IgG2 | 12 pairs | 4 pairs (linked through the hinge region) | Heavy chain: hinge region; Light chain and heavy chain: Light chain's terminal cysteine and heavy chain's third cysteine |
| IgG3 | 12 pairs | 11 pairs (linked through the hinge region) | Heavy chain: hinge region; Light chain and heavy chain: Light chain's terminal cysteine and heavy chain's third cysteine |
| IgG4 | 12 pairs | 2 pairs (linked through the hinge region) | Heavy chain: hinge region; Light chain and heavy chain: Light chain's terminal cysteine and heavy chain's third cysteine |
Impact of Disulfide Bonds on Antibody Function
The functional properties of immunoglobulin G (IgG) antibody subclasses are significantly affected by the number and positioning of disulfide bonds. Notably, the number of interchain disulfide bonds differs among these subclasses: IgG1 and IgG4 each feature 2 pairs, IgG2 comprises 4 pairs, while IgG3 is characterized by an extensive 11 pairs. This variation has a direct impact on the structural stability and rigidity of the antibodies. An increased number of interchain disulfide bonds enhances structural stability, which is critical for sustaining prolonged immune responses. For instance, the elevated number of interchain disulfide bonds in IgG3 may confer increased stability and efficacy in neutralizing specific pathogens. Furthermore, the specific location of disulfide bonds between the light and heavy chains affects the binding angles and affinity of antibodies for antigens. These structural variations influence the antibodies' antigen recognition and binding capabilities, conferring each subclass with unique functional attributes necessary to address the organism's diverse immunological defense requirements.
Disulfide Bonds and Antibody Conformational Flexibility
The hydrophobic characteristics of both intrachain and interchain disulfide bonds play a crucial role in determining the conformational flexibility of antibodies. Intrachain disulfide bonds primarily contribute to the stabilization of local peptide segments within individual chains, thereby constraining conformational alterations in these specific regions. Conversely, interchain disulfide bonds, which connect distinct peptide chains, exert a more pronounced effect on the overall rigidity and flexibility of the antibody structure. In IgG1, for instance, the cysteine residues that participate in forming interchain disulfide bonds are situated in the hinge region. This positioning imparts greater hinge flexibility when compared to IgG2, IgG3, and IgG4, where interchain disulfide bonds occur between cysteine residues located at the junction of the VH and CH1 domains, resulting in a more rigid conformation. The differential hydrophobic interactions and their effects on the antibody's comprehensive structure lead to varying degrees of flexibility during antigen binding and conformational changes throughout immune response activation. Such flexibility is essential for the antibody's biological activity and its capacity to effectively mediate immunological functions.
Formation and Regulation of Disulfide Bonds
Disulfide Bond Formation in Mammalian Cells
In the biological world, the formation of disulfide bonds in intracellular proteins differs between prokaryotes and eukaryotes, although both share similar underlying mechanisms. Prokaryotic organisms rely on a series of oxidoreductases to catalyze the formation of disulfide bonds, while eukaryotic organisms depend on a set of protein disulfide isomerases (PDIs). Despite these differences, both systems employ an electron transfer chain to drive reactions, involving oxidation, reduction, and isomerization.
In mammalian cells, when a polypeptide is translocated into the endoplasmic reticulum (ER) through the Sec61 translocon, it associates with PDIs, which facilitate the introduction of disulfide bonds through redox reactions. Disulfide bonds that have already formed on the substrate protein can also be reduced, thereby enabling the formation of new disulfide linkages. These oxidation, reduction, and isomerization reactions may occur through a two-step or one-step mechanism. In the two-step process, two enzymes are involved: an isomerase reduces the disulfide bond, and an oxidase forms a new one. In the one-step mechanism, only one enzyme from the PDI family—acting as both the oxidase and reductase—is involved. These reactions dynamically regulate protein function, ensuring that nascent polypeptides fold correctly, allowing for the maturation of post-translational modifications and the subsequent secretion of fully processed proteins.
Enzymes Involved in Disulfide Bond Formation
A variety of enzymes are involved in the formation of disulfide bonds, with protein disulfide isomerase (PDI) being a central player. The PDI family comprises at least 15 members that facilitate disulfide bond exchange. These enzymes contain at least one catalytic cysteine residue with a CXXC motif, a structural feature that enables them to switch between oxidized and reduced states.
The PDI family functions as a key component in the electron transfer chain between nascent polypeptides and oxidases during disulfide bond formation. Specifically, PDI enzymes catalyze the flow of electrons from donor molecules such as glutathione, promoting the reduction of disulfide bonds. During the isomerization process, oxidoreductases attack pre-existing disulfide bonds, forming mixed disulfides, before reducing the free sulfhydryl groups and re-establishing the correct disulfide linkages. Due to their relatively unstable disulfide-active sites, PDI family members can act as electron acceptors, facilitating conformational changes that enable diverse disulfide bond rearrangements. This ensures the correct formation of disulfide bonds, which are essential for the structural integrity and functionality of the protein.
Regulatory Mechanisms of Disulfide Bond Formation
The formation of disulfide bonds is meticulously regulated by a variety of factors. A principal regulatory element is the cellular redox state, which profoundly influences the enzymatic activity involved in disulfide bond formation. Specifically, an oxidizing intracellular environment facilitates the formation of disulfide bonds, whereas a reducing environment may reverse the process.
Furthermore, the microenvironment within the endoplasmic reticulum (ER) plays an indispensable role in modulating disulfide bond formation. Key factors such as the concentration of calcium ions and the pH within the ER can affect both the activity and conformation of protein disulfide isomerases (PDIs) and other associated enzymes, thereby indirectly regulating the disulfide bond formation process.
Additionally, cellular signaling pathways contribute significantly to this regulation. Specific signaling molecules can either activate or inhibit the expression of genes involved in disulfide bond formation, thus influencing the synthesis of the relevant enzymes. This precisely coordinated regulation ensures that proteins fold correctly and achieve their functional conformations, which is essential for their biological activity.
Reduction of Disulfide Bonds and Influencing Factors
Disulfide Bond Reduction Reaction
Disulfide bonds are susceptible to cleavage in the presence of reducing agents. In biochemical practices, thiol compounds such as β-mercaptoethanol (β-ME) and dithiothreitol (DTT) are commonly utilized as reductants. Achieving complete cleavage of disulfide bonds typically necessitates an excess of the thiol reagent. Additionally, tris(2-carboxyethyl)phosphine (TCEP) serves as an effective reductant. In contrast to β-ME and DTT, TCEP is odorless, selective, and functional across both alkaline and acidic conditions. It is also more hydrophilic and resistant to oxidation, allowing it generally to remain present without removal prior to protein thiol modification.
The primary mechanism underlying disulfide bond formation and rearrangement in proteins is the redox exchange reaction between thiols and disulfide bonds. The rearrangement of disulfide bonds within proteins is mediated through thiol-disulfide exchange reactions. During this process, a thiol group from a cysteine residue engages with an existing disulfide bond within the same protein, a phenomenon referred to as "disulfide shuffling." Although this process does not alter the total number of disulfide bonds, it does modify their positions within the protein structure.
Mechanism of Monoclonal Antibody Disulfide Bond Reduction
Reduction of disulfide bonds in monoclonal antibodies predominantly involves redox reactions orchestrated by oxidoreductases. The glutathione and thioredoxin systems, which encompass thioredoxin (Trx), thioredoxin reductase (TrxR), and nicotinamide adenine dinucleotide phosphate (NADPH), are well-established enzymatic mechanisms facilitating this reduction process.
In this context, NADPH, produced via the pentose phosphate pathway, acts as the primary electron donor for disulfide bond reduction. The transfer of electrons proceeds from NADPH to TrxR, leading to the reduction of disulfide bonds within TrxR. Thereafter, these electrons are conveyed to oxidized thioredoxin (Trx), thereby converting it into its reduced state. Subsequently, reduced thioredoxin catalyzes the reduction of disulfide bonds present in antibodies. Analogous to the thioredoxin system, glutathione (GSH) operates in a similar fashion. Together, these systems synergistically contribute to the reduction of disulfide bonds in monoclonal antibodies. They play a pivotal role in modulating intracellular redox homeostasis and ensuring optimal protein functionality.
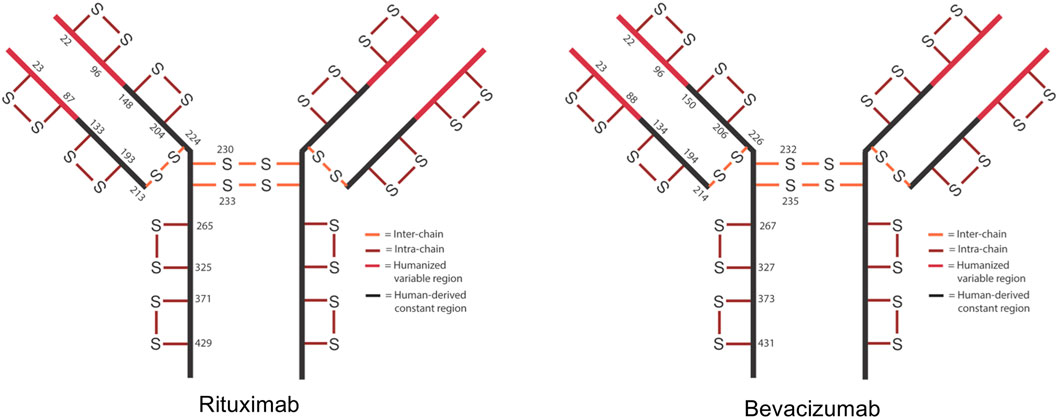 Schematic of expected disulfide bond locations for rituximab and bevacizumab.
Schematic of expected disulfide bond locations for rituximab and bevacizumab.Factors Affecting Disulfide Bond Reduction
In the production of antibody-based therapeutics, numerous parameters have the potential to impact the reduction of disulfide bonds. A prominent factor is the applied shear force during cell harvesting. Elevated shear forces can lead to increased cell lysis, consequently releasing proteases engaged in redox reactions such as thioredoxin reductase and glutathione reductase, along with relevant cofactors like NADPH. These events may precipitate the reduction of disulfide bonds in the therapeutic protein.
Oxygen availability constitutes another influential element. Insufficient levels of dissolved oxygen in the cell harvest medium can lead to inadequate NADPH consumption, thereby fostering conditions conducive to disulfide bond reduction. Regarding pH levels, maintaining an environment near neutrality promotes the oxidation of NADPH, rendering the harvest solution more susceptible to disulfide bond reduction.
Temperature also plays a crucial role. Lower storage temperatures tend to decelerate redox reaction rates, potentially diminishing the incidence of disulfide bond reduction. Moreover, the intracellular redox state exerts a significant influence on disulfide bond reduction. Variations in the oxidative or reductive milieu within cells can directly alter the activity of the enzymes involved, thereby affecting both the rate and the extent of disulfide bond reduction.
Prevention and Characterization of Disulfide Bonds
1. Fundamental Strategies for Preventing Disulfide Bond Reduction
To mitigate or prevent the reduction of disulfide bonds in antibody molecules during production, various strategies can be employed. One common approach is the addition of inhibitors to obstruct the expression of relevant reductases, effectively preventing or reducing disulfide bond reduction. It is crucial, however, to ensure that the inclusion of such inhibitors does not adversely affect cell growth or the expression of target proteins. In scenarios where multiple reductases are implicated in disulfide bond reduction, the introduction of diverse protease expression inhibitors might be warranted.
Regulating process parameters plays a pivotal role as well. For instance, the addition of EDTA or metal ions can inhibit the activity of related reductases. However, it is essential to determine optimal timing for their introduction during the process and to develop methods for their effective removal afterward, to prevent adverse impacts on product quality. Enhancing dissolved oxygen levels, introducing hydrogen peroxide, and employing molecules like L-cysteine, which can interact and deplete key reductases, may further aid in preventing or reducing disulfide bond reduction.
Furthermore, minimizing shear forces during the cell harvest stage can reduce the release of these reductases. Adjusting the pH of the cell harvest fluid and lowering storage temperatures can also delay disulfide bond reduction. When employing these strategies, it is important to ensure that the molecular quality of the product is not compromised by pH and temperature adjustments.
2. Methods for Characterizing Disulfide Bonds
Characterization of disulfide bonds is typically conducted through mass spectrometry of enzymatically digested peptides. By assessing the molecular weights of digests under reduced and non-reduced conditions, or through secondary mass spectrometry fragments of disulfide-bonded peptides, the pairing of antibody disulfide bonds can be confirmed. This method is especially suitable for IgG-type antibodies, which are assembled through interchain disulfide bonds between light and heavy chains.
In practical applications, antibodies are subjected to enzymatic digestion and analyzed separately under reduced and non-reduced conditions. Comparing the molecular weights of these digests under different conditions, in conjunction with secondary mass spectrometry fragment information, offers clear insights into disulfide bond pairing. This approach yields precise information regarding disulfide bonds, providing a critical foundation for subsequent research and production.
Select Service
Learn More
3. The Importance of Disulfide Bond Characterization
Characterizing disulfide bonds is of utmost significance. The number and location of disulfide bonds are crucial quality attributes of monoclonal antibodies (CQAs); improper formation can lead to loss of biological activity or provoke host immune responses.
In the production of therapeutic proteins, due to process variation and the inherent complexity of the products, numerous CQAs related to both the process and the product itself require vigilant monitoring, with disulfide bonds being among these essential attributes. Accurate characterization of disulfide bonds ensures that the drug maintains the correct structure and function, thereby guaranteeing its quality. Only when a drug meets the quality standards can it achieve its expected therapeutic effect in clinical applications and ensure patient treatment efficacy. Additionally, proper disulfide bond structures help prevent immune responses triggered by structural anomalies, ensuring drug safety and safeguarding patient well-being.
The Role of Disulfide Bonds in Protein Activity and Monoclonal Antibody Stability
1. Impact of Disulfide Bonds on Protein Activity
Disulfide bonds play a multifaceted and critical role in preserving protein activity. Primarily, they constitute significant components of active sites in enzymes, a feature that is particularly pronounced in certain oxidoreductases. For instance, disulfide bond oxidoreductases (Dsb) and protein disulfide isomerase (PDI) leverage these bonds to catalyze oxidation, reduction, and isomerization reactions efficiently, thereby ensuring the seamless progression of enzymatic reactions and maintaining protein metabolism and functionality.
Furthermore, through their dynamic process of breaking and forming, disulfide bonds can induce conformational changes in proteins via allosteric effects, thus finely tuning protein function. Taking the natriuretic peptide receptor-A protein as an example, the formation of a dimeric structure through inter-molecular disulfide bonds is essential for activating its biological function. Additionally, certain proteins require the formation of all natural disulfide bonds to fold into their correct conformations, as exemplified by bovine pancreatic ribonuclease A, proinsulin, and the bovine pancreatic trypsin inhibitor. Since protein folding invariably accompanies the formation and transformation of disulfide bonds, they can serve as markers for in-depth exploration of protein folding pathways, thereby providing vital insights into the functional mechanisms of proteins.
2. Influence of Disulfide Bond Reduction on Monoclonal Antibody Stability in Downstream Processing
During downstream processing, mismatches or reductions in disulfide bonds can differentially impact monoclonal antibody stability. In the Protein-A affinity chromatography step, monoclonal antibodies primarily engage in specific interactions with Protein-A ligands via the Fc region. Research reveals that monoclonal antibodies with reduced disulfide bonds retain their higher-order structure, meaning their affinity for Protein-A resin remains unchanged. Consequently, disulfide bond reduction does not significantly affect this step.
In contrast, the low pH viral inactivation phase presents more complexity. Studies indicate that reductions in disulfide bonds following low pH treatment can enhance protein aggregation; however, results vary, and this effect may not be consistent, suggesting that aggregation might also be influenced by molecular intrinsic properties like type and hydrophobicity.
In ion exchange chromatography, the reduction of disulfide bonds may alter the surface charge distribution of antibodies, potentially leading to anomalous peaks in CEX-HPLC and AEX-HPLC analyses. Comparisons of monoclonal antibody samples with varying reduction levels reveal that while the overall peak shapes in AEX and CEX resemble each other, more pronounced tailing peaks appear during AEX capture and CEX elution in samples with higher reduction levels. Nevertheless, it remains uncertain whether these chromatographic differences are universally applicable. Overall, the subtle differences in chromatograms make it challenging to identify reduced disulfide bonds or determine reduction levels based on these results alone. Thus, it is imperative to employ rapid and precise analytical methods to monitor disulfide bond reduction during downstream purification to ensure the stability and quality of monoclonal antibodies.
References
- Laitaoja M, Tossavainen H, Pihlajamaa T, et al. Redox-dependent disulfide bond formation in SAP30L corepressor protein: Implications for structure and function. Protein Science. 2016;25(2):487-498. DOI: 10.1002/pro.2849
- O'Mara B, Gao ZH, Kuruganti M, Mallett R, Nayar G, Smith L, Meyer JD, Therriault J, Miller C, Cisney J, Fann J. Impact of depth filtration on disulfide bond reduction during downstream processing of monoclonal antibodies from CHO cell cultures. Biotechnol Bioeng. 2019 Jul;116(7):1669-1683. DOI: 10.1002/bit.26964. Epub 2019 Apr 4. PMID: 30883673.
- Tan Z, Ehamparanathan V, Ren T, et al. On-column disulfide bond formation of monoclonal antibodies during Protein A chromatography eliminates low molecular weight species and rescues reduced antibodies. mAbs. 2020;12(1):e1829333. DOI: 10.1080/19420862.2020.1829333
- Guo J, Carta G. Unfolding and aggregation of a glycosylated monoclonal antibody on a cation exchange column. Part II. Protein structure effects by hydrogen deuterium exchange mass spectrometry. J Chromatogr A. 2014 Aug 22;1356:129-37. DOI: 10.1016/j.chroma.2014.06.038. Epub 2014 Jun 19. PMID: 25011681.
- Merdanovic M, Mogk A, Tomoyasu T, et al. Disulfide bond formation and redox regulation in the Golgi apparatus. FEBS Letters. 2022;596(22):2859-2872. DOI: 10.1002/1873-3468.14510
- Sevier, C., Kaiser, C. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol 3, 836–847 (2002). https://doi.org/10.1038/nrm954
- Hatahet F, Ruddock LW. Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxidants & Redox Signaling. 2009;11(11):2807-2850. DOI: 10.1089/ars.2009.2466
- Chaturvedi S, Bawake S, Sharma N. Recent advancements in disulfide bridge characterization: Insights from mass spectrometry. Rapid Commun Mass Spectrom. 2024 Apr 15;38(7):e9713. DOI: 10.1002/rcm.9713. PMID: 38361473.











