Biomarker Identification Analysis
Why Biomarker Identification is Important?
Biomarkers are measurable biological indicators that reflect physiological states, pathological processes, or responses to interventions. They provide essential insights into molecular events and disease mechanisms. Accurate biomarker identification is pivotal for life science research and translational studies. Biomarkers can span nucleic acids, proteins, metabolites, epigenetic modifications, and cellular components.
Reliable biomarkers are characterized by specificity and reproducibility. They often correlate with disease progression or distinct biological phenotypes. In the context of precision research, biomarkers guide the selection of targets for molecular studies, stratify biological samples, and enhance understanding of complex molecular networks.
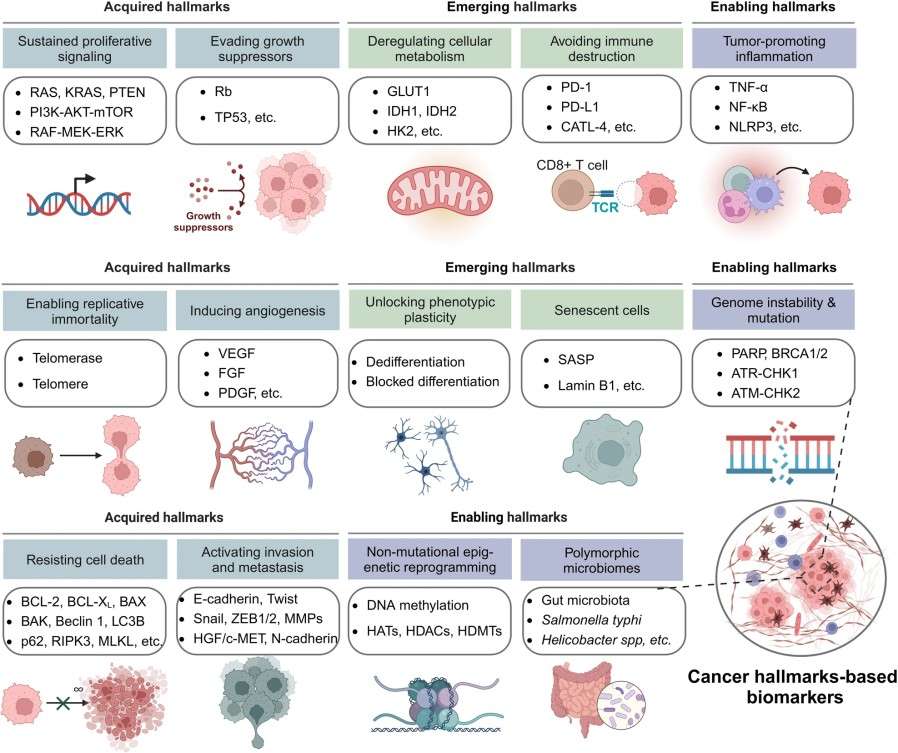
Figure 1. The 14 cancer hallmarks-based biomarkers (Zhou Y, et al., 2024).
MS-Based Proteomics for Biomarker Identification
High-Resolution Protein Profiling
Mass spectrometry enables researchers to simultaneously measure the abundance, structure, and modifications of thousands of proteins. High-resolution instruments generate precise and reproducible protein data. These tools can detect even proteins present at very low levels, which are often the most important biomarkers.
Quantitative Accuracy
Quantitative proteomics strategies enable the accurate comparison of protein levels across different conditions or patient groups. Methods like label-free quantification or stable isotope labeling provide clear numerical values for protein abundance.
Detection of PTMs
Proteins often undergo chemical changes after they are produced, such as phosphorylation or glycosylation. Mass spectrometry can detect these modifications, revealing functional changes that may indicate disease processes.
Quantitative Proteomics Strategies for Biomarker Discovery
Quantitative proteomics provides the foundation for identifying reliable biomarkers by accurately measuring protein abundance across biological samples.
- Label-free quantification measures protein abundance directly from the intensity of peptide ions detected by mass spectrometry. Spectral counting estimates protein abundance by counting the number of MS/MS spectra assigned to peptides of a given protein.
- SILAC incorporates heavy isotope-labeled amino acids into the proteome of cultured cells. After mixing labeled and unlabeled samples, MS analysis quantifies relative protein abundance based on peptide ion intensity ratios. SILAC offers high accuracy and low technical variability, making it an ideal tool for mechanistic studies in cell culture.
- TMT and iTRAQ are chemical labeling strategies applied to peptides after protein digestion. These isobaric tags allow multiplexing of multiple samples in a single MS run, increasing throughput and experimental consistency.
Integrating Multi-Omics to Improve Biomarker Specificity
Multi-omics integration combines complementary layers of molecular information, thereby improving the specificity and robustness of biomarker discovery. Key multi-omics modalities include
- Genomics: Analyzing changes at the DNA level, including mutations, gene copy number gains or losses, and single-base changes, helps reveal both inherited and acquired factors that contribute to disease. Connecting these genetic changes to their effects on gene activity or protein function makes biomarkers more meaningful and biologically relevant.
- Transcriptomics: RNA-level profiling reveals dynamic gene expression patterns, alternative splicing events, and non-coding RNA activity. RNA sequencing captures transcriptional changes resulting from genetic or environmental perturbations.
- Proteomics: MS-based proteomics enables precise measurement of protein levels, chemical modifications, and protein interactions. When combined with transcriptomic data, it helps confirm whether changes seen at the gene expression level result in meaningful changes in protein function.
- Metabolomics: Cellular metabolites show the real-time metabolic state of cells and tissues. When metabolomic data are combined with proteomic profiles, enzyme activity can be directly connected to functional biological changes. This integrated view helps identify biomarkers that capture both upstream molecular regulation and downstream physiological effects.
Creative Proteomics provides integrated analysis:
Integrative Analysis of PTMs and Metabolomics
Integrative Analysis Services of Proteomics and Phosphoproteome
Integrative Analysis Services of Proteomics-Acetyl
Validation Strategies for Biomarkers
Validation ensures that the biomarkers are robust indicators of the intended biological state and suitable for downstream applications.
- Parallel Reaction Monitoring (PRM): PRM provides precise and sensitive quantification of selected peptides in complex biological matrices. PRM focuses on predefined targets that minimize background interference, increase reproducibility, and allow accurate measurement of low-abundance proteins.
- ELISA: It is a highly sensitive and specific method for validating biomarker candidates. It can measure proteins in various biological fluids and is well-suited for studies that track changes over time across large groups of samples.
- Antibody generation and assay development: High-affinity monoclonal or polyclonal antibodies can be designed against specific protein epitopes. These antibodies facilitate multiple downstream applications, including WB, IHC, flow cytometry, and custom assay platforms.
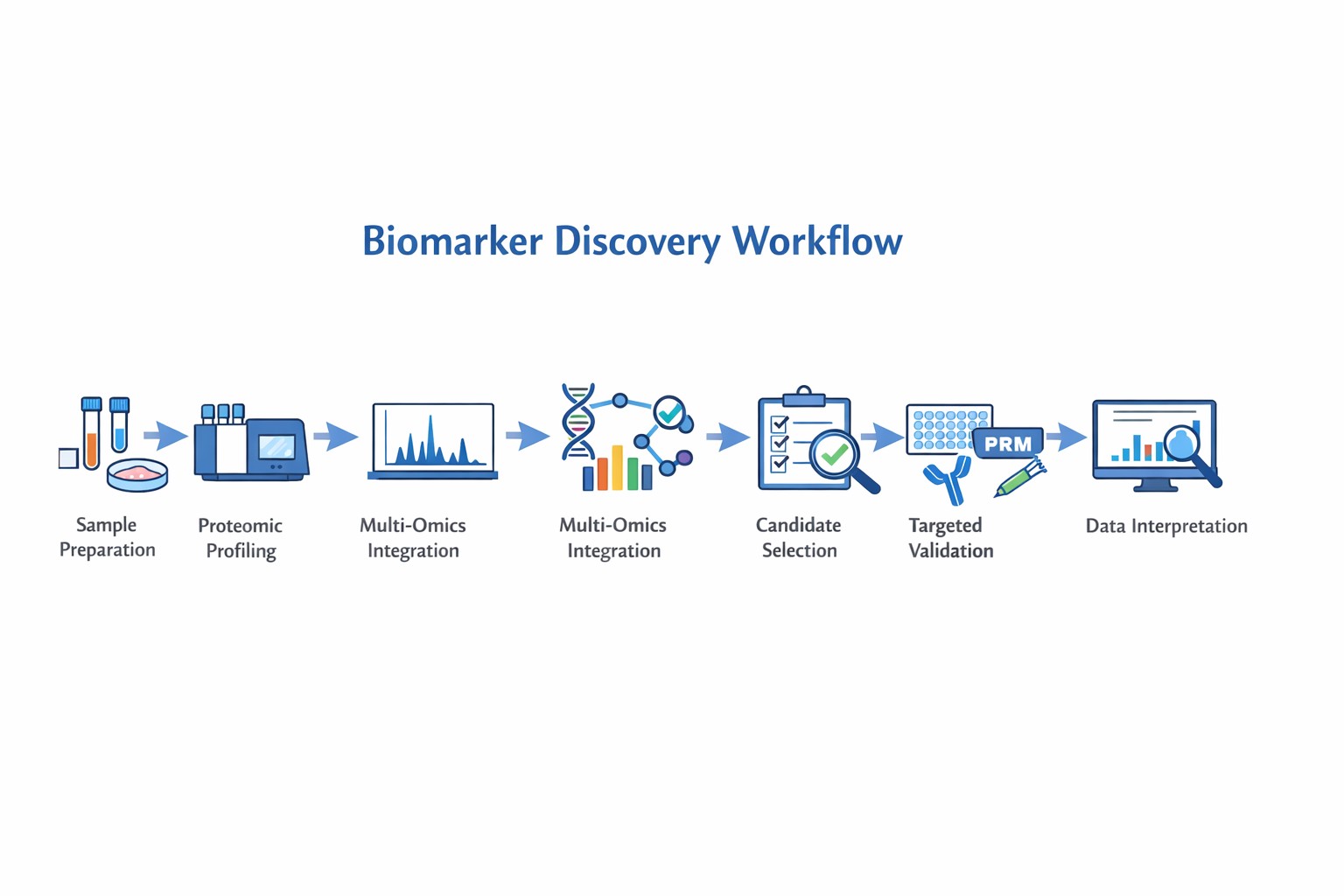
Our Biomarker Discovery Workflow
- Sample Preparation: Collection, lysis, and protein extraction from diverse biological samples.
- Proteomic Profiling: High-resolution MS analysis, peptide separation, and quantification.
- Multi-Omics Integration: Correlation with genomic, transcriptomic, metabolomic, and epigenomic data.
- Candidate Selection: Statistical and bioinformatics filtering to identify potential biomarkers.
- Targeted Validation: PRM, ELISA, and antibody-based assays to confirm specificity.
- Data Interpretation: Comprehensive bioinformatic analysis to support biological conclusions.
Data Deliverables & What You Receive
- Quantitative proteomic datasets with protein abundance and modification profiles.
- Integrated multi-omics analyses highlighting candidate biomarkers.
- Statistical validation reports and confidence scores.
- Targeted assay results, including PRM or ELISA confirmation.
- Bioinformatics interpretations with pathway mapping and functional annotations.
Applications Across Life Science and Translational Research
- Elucidating disease mechanisms and molecular pathways.
- Stratifying biological samples in cohort studies.
- Identifying predictive and pharmacodynamic markers.
- Supporting clinical sample analysis and experimental design.
- Enhancing translational research through mechanistic insight.
Sample Requirements
| Sample Type | Recommended Format | Notes |
| Tissue | Fresh, frozen, or FFPE | Minimum quantity depends on tissue type. |
| Blood/Plasma/Serum | EDTA, heparin, or citrate collection | Ensure proper storage to preserve protein integrity. |
| Cell Lines | Whole cells or lysates | Harvest at exponential growth phase. |
| Biofluids | Urine, cerebrospinal fluid, saliva | Pre-treatment may be required. |
| Experimental Models | Homogenates, organoids, xenografts | Standardized collection for reproducibility. |
Why Choose Creative Proteomics
- Technical Expertise: Extensive experience in biomarker identification and multi-omics analysis.
- Advanced Instrumentation: Access to high-resolution mass spectrometry platforms and robust bioinformatics tools.
- Multi-Omics Integration: Capability to combine proteomics with genomics, transcriptomics, metabolomics, and epigenomics for comprehensive analysis.
- Precision and Reproducibility: Emphasis on reliable identification and validation of candidate biomarkers.
FAQ
-
Q1: Why do some biomarker candidates fail validation or translation?
A1: Many biomarker candidates discovered in early studies do not hold up in later work. Common reasons include small or unbalanced sample sets, inconsistent data across experiments, and the absence of independent confirmation. To overcome these limitations, robust statistical analysis, testing in separate sample groups, and validation using complementary methods are essential.
-
Q2: Can multi-omics analysis identify low-abundance biomarkers?
A2: Yes. Combining data from genomics, transcriptomics, proteomics, and metabolomics enables the detection of small but biologically significant changes, even when the molecules of interest are present at very low levels.
-
Q3: Can biomarker discovery identify dynamic or temporal changes in disease progression?
A3: Yes. Collecting samples over time and analyzing them using quantitative proteomics and integrated multi-omics enables the tracking of molecular changes as they occur. This approach helps identify dynamic biomarkers that reflect disease progression or biological responses to experimental interventions.
Demo
Demo: LC‑MS/MS Based Metabolomics and Proteomics Reveal Candidate Biomarkers and Molecular Mechanism of Early IgA Nephropathy
This integrative study combined untargeted LC-MS/MS proteomics and metabolomics to investigate plasma samples from patients with IgA nephropathy versus those from healthy controls. Differentially expressed proteins and metabolites were identified and analyzed with machine learning. Key candidates such as PRKAR2A, IL6ST, and SOS1 were highlighted. Independent validation showed strong classification performance, underscoring the value of combined omics for early biomarker discovery.
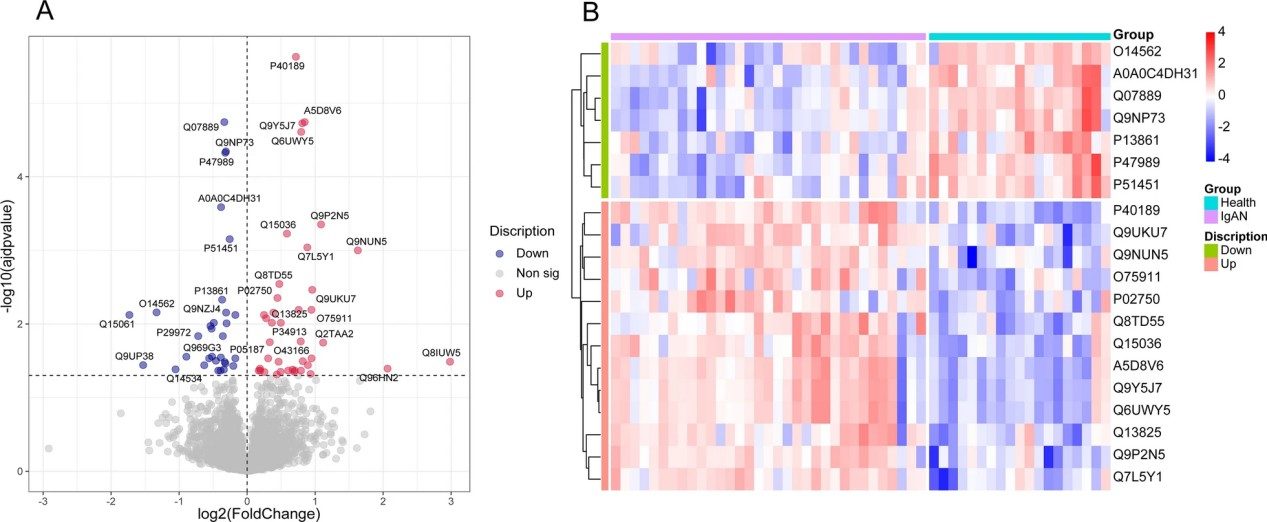
Figure 2. Proteomic analysis of IgAN (Zhang D, et al., 2022).
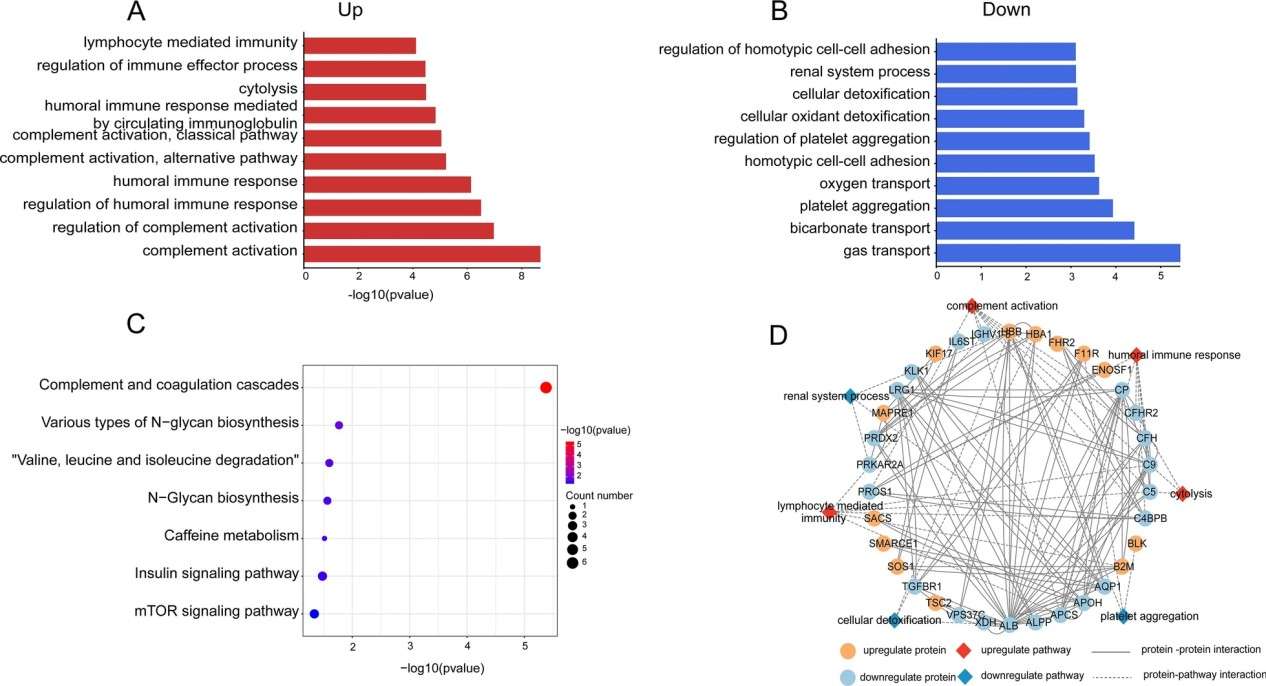
Figure 3. GO terms and KEGG pathway analysis for DEPs (Zhang D, et al., 2022).
-
Case Study
Case: Discovery of Novel Biomarkers for Diagnosing and Predicting the Progression of Multiple Sclerosis Using TMT‑Based Quantitative Proteomics
Abstract
Multiple sclerosis (MS) is a chronic inflammatory and neurodegenerative condition of the central nervous system. Biomarker discovery for MS is challenging due to disease complexity and overlapping clinical features with other neurological disorders. Reliable protein biomarkers in cerebrospinal fluid (CSF) and plasma can improve molecular insights and support stratification of disease states.
The study aimed to identify protein biomarkers that differentiate MS patients from non‑inflammatory neurological controls using quantitative proteomics. The authors also sought to evaluate whether candidate proteins could distinguish disease progression phenotypes.
Methods
- Employed an isobaric mass tag labeling strategy (Tandem Mass Tag, TMT) coupled with high‑resolution mass spectrometry to quantify protein expression.
- Differentially expressed proteins (DEPs) were identified through proteomic bioinformatics.
- Candidate proteins were further validated in a larger cohort of 160 samples (paired CSF and plasma) using enzyme‑linked immunoassay (ELISA).
- Receiver operating characteristic (ROC) curves assessed diagnostic relevance.
Results
- Out of 343 quantified proteins, 83 were differentially expressed between MS patients and controls.
- Functional enrichment analysis revealed involvement in processes such as platelet degranulation and signaling pathways relevant to immune regulation.
- Three proteins emerged as central in the protein–protein interaction network.
- ELISA validation confirmed significant upregulation of IGFBP7 and downregulation of SST in CSF from MS patients.
- IGFBP7 also demonstrated potential to discriminate between relapsing‑remitting and secondary progressive MS phenotypes.
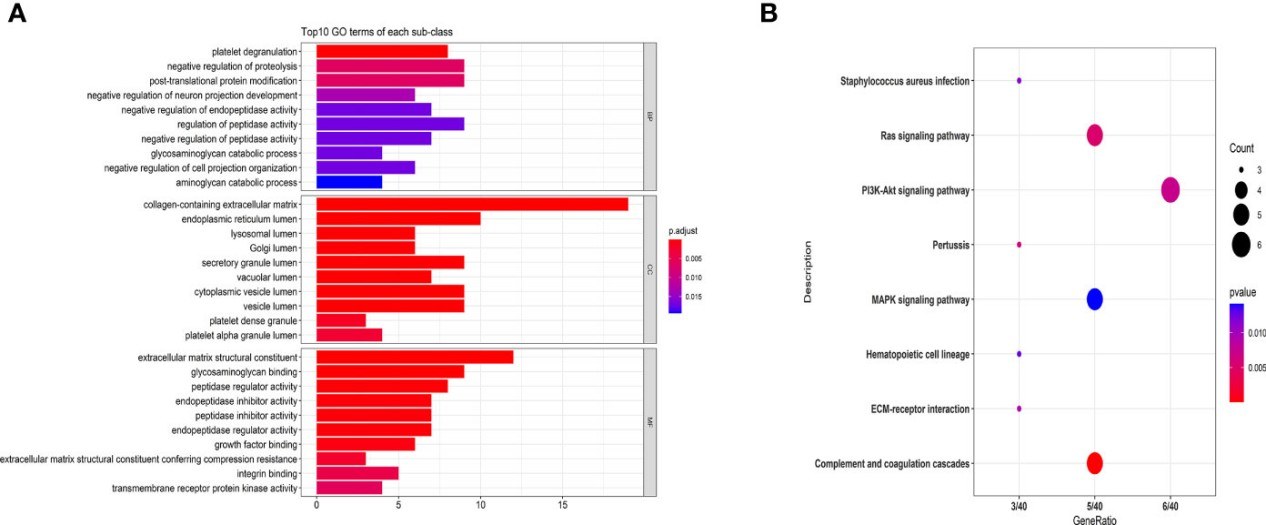
Figure 4. GO and KEGG pathway analyses of MS-related proteins.
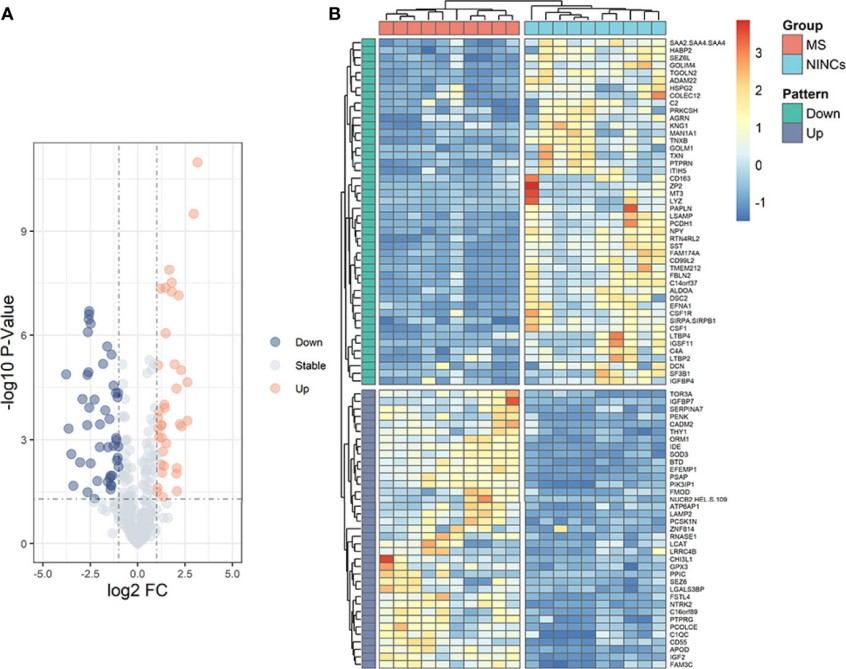
Figure 5. Volcano plot and heatmap of patients with MS vs NINCs.
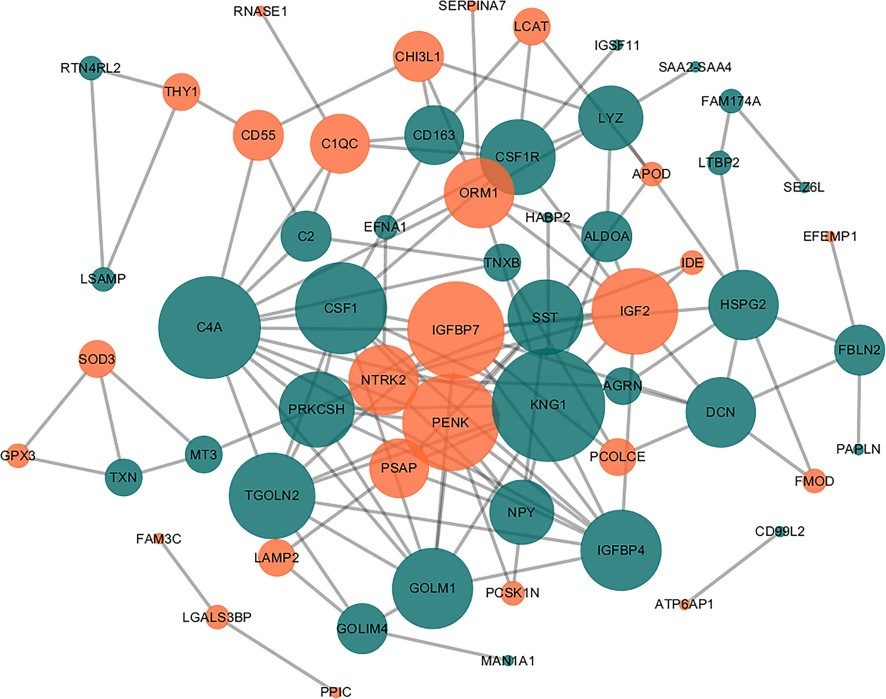
Figure 6. Interaction network of the DEPs.
Conclusion
High-throughput quantitative proteomics, coupled with TMT labeling, can reveal robust protein biomarkers in complex clinical samples. IGFBP7 and SST show promise as protein indicators associated with MS pathology, with IGFBP7 potentially serving as a key marker for disease classification and progression.
Related Services
References
- Zhou Y, et al. Tumor biomarkers for diagnosis, prognosis and targeted therapy. Signal transduction and targeted therapy, 2024, 9(1): 132.
- Zhang D, et al. LC-MS/MS based metabolomics and proteomics reveal candidate biomarkers and molecular mechanism of early IgA nephropathy. Clinical proteomics, 2022, 19(1): 51.
- Shi Y, et al. Discovery of novel biomarkers for diagnosing and predicting the progression of multiple sclerosis using TMT-based quantitative proteomics. Frontiers in Immunology, 2021, 12: 700031.