Cysteine-Targeted Covalent Drugs Analysis
What is cysteine used for?
Covalent small molecules have gained renewed attention due to their ability to form stable, irreversible interactions with specific amino acid residues in proteins. Among these residues, cysteine stands out because of its unique chemical reactivity and relatively low abundance in the proteome. This combination enables selective targeting while minimizing widespread nonspecific binding.
From a proteomics perspective, understanding how small molecules interact with cysteine residues requires quantitative, site-resolved analytical strategies. Advances in mass spectrometry-based proteomics and chemoproteomics now allow systematic mapping of cysteine engagement across complex biological systems, providing critical insights into molecular selectivity and protein interaction landscapes.
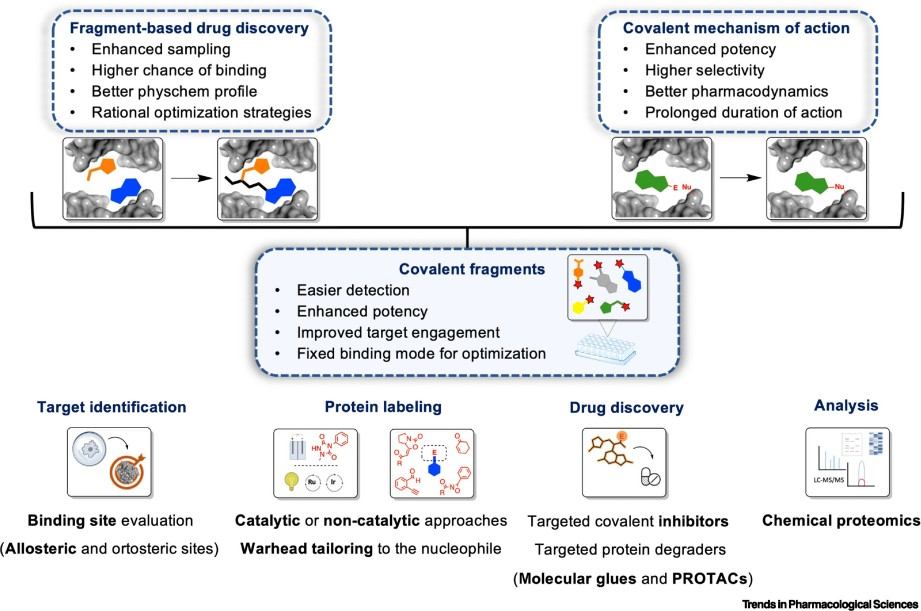
Figure 1. The concept of covalent fragments, their warheads and labeling chemistries (Csorba N, et al., 2023).
Chemical and Structural Characteristics of Cysteine
Cysteine contains a nucleophilic thiol (–SH) side chain that can participate in redox reactions, metal coordination, and covalent modification. In proteins, cysteine residues may be solvent-exposed, buried within folded structures, or involved in disulfide bonds, leading to diverse reactivity profiles.
This heterogeneity is a key reason why cysteine-targeted covalent interactions are highly context-dependent. Quantitative proteomics approaches are therefore essential to distinguish reactive, ligandable cysteines from those that are structurally inaccessible or functionally constrained.
Principles of Cysteine-Targeted Covalent Binding
Cysteine-targeted covalent molecules typically rely on electrophilic functional groups that react with thiol groups under physiological conditions. The binding process involves two conceptual steps:
- Recognition and positioning, where the molecule associates with the protein through non-covalent interactions
- Covalent bond formation, where the electrophile reacts with the cysteine thiol
Quantitative analysis of this process focuses on occupancy, selectivity, and competition at the residue level. These parameters cannot be reliably inferred from bulk assays alone and require proteome-wide measurement strategies.
Chemoproteomics Strategies for Cysteine-Targeted Covalent Drugs
Chemoproteomics integrates chemical probes with mass spectrometry to profile protein-small molecule interactions directly in complex samples. For cysteine-focused studies, thiol-reactive probes are commonly used to label accessible cysteine residues across the proteome.
When a covalent small molecule occupies a cysteine site, probe labeling at that position is reduced. By quantitatively comparing probe signals between treated and control samples, researchers can infer which cysteine residues are engaged and to what extent. This strategy enables unbiased identification of both primary interaction partners and secondary binding proteins.
Why Identifying Cysteine Binding Sites Is Critical
Knowing which cysteine residue is modified is as important as identifying the target protein itself. Site-specific information enables researchers to:
- Distinguish functional binding from nonspecific reactivity
- Compare binding patterns across compound analogs
- Support structure–activity relationship optimization
- Evaluate selectivity across protein families
Proteomics-based cysteine mapping provides this information directly at the peptide and residue level, offering clarity that cannot be achieved through bulk readouts alone.
Quantitative Proteomics for Occupancy and Selectivity Assessment
Quantitative proteomics enables comparison of cysteine labeling intensities across experimental conditions, providing insight into binding occupancy and selectivity. By integrating labeling strategies with high-resolution LC-MS/MS, researchers can determine:
- Relative occupancy at individual cysteine sites
- Dose-dependent engagement patterns
- Competitive displacement across compounds
Creative Proteomics' Service Workflow
- Compound incubation in biological systems
- Labeling with cysteine-reactive probes
- Affinity enrichment of modified peptides
- High-resolution LC-MS/MS analysis
- Bioinformatics-driven site identification and quantification

Applications in Molecular Research and Lead Optimization
- Identification of ligandable cysteine residues
- Comparative profiling of structurally related compounds
- Mechanistic studies of protein–small molecule interactions
- Early assessment of proteome-wide reactivity trends
Sample Requirements
| Item | Quick guidance |
| Compatible sample types | Purified protein, cell lysate, intact cells, fresh/frozen tissue. |
| Recommended input | Purified protein: 1–10 µg; Lysate/cells: 100–1,000 µg total protein; Tissue: ≥1 mg wet weight (platform-dependent). |
| Redox & handling | Minimize air/oxidation. |
| Controls & replication | Vehicle control, probe-only, competition (drug+probe); ≥3 biological replicates recommended. |
| Storage & transport | Snap-freeze samples, store at −80°C, avoid repeated freeze–thaw cycles. |
| Special samples (FFPE/tissues) | Require deparaffinization and antigen retrieval. |
Why Choose Creative Proteomics for Cysteine-Targeted Covalent Drug Analysis
- Expertise & Experience: Over years in proteomics and chemoproteomics, specializing in cysteine-targeted analysis.
- Comprehensive Workflow: From probe design to mass spectrometry and bioinformatics interpretation.
- Advanced Instrumentation: State-of-the-art Orbitrap and timsTOF platforms for high-resolution, site-specific data.
- High Specificity & Depth: Detects cysteine engagement with minimal off-target effects across diverse sample types.
- Reliable & Reproducible Results: Rigorous quality management ensures consistent, actionable data.
FAQ
-
Q1: Can chemoproteomic methods quantify how strongly a covalent ligand engages a cysteine site?
A1: Yes. Competitive chemoproteomic strategies compare probe labeling intensities across multiple ligand concentrations to derive metrics such as occupancy and dose-response, allowing for a quantitative assessment of how effectively a ligand engages a cysteine site under defined conditions.
-
Q2: Do covalent small molecules exclusively react with cysteine residues?
A2: Although cysteine is a common target due to its nucleophilicity, other residues such as lysine, serine, and tyrosine can also undergo covalent modification depending on the electrophile used. Mapping of ligandability across the proteome shows the importance of profiling multiple residue types for comprehensive interaction landscapes.
-
Q3: How do reversible and irreversible covalent inhibitors differ in cysteine targeting?
A3: Irreversible inhibitors form permanent covalent bonds with cysteine residues, often leading to prolonged site occupancy. Reversible covalent inhibitors form bonds that can dissociate under physiological conditions, offering the potential for higher selectivity and reduced off-target toxicity.
-
Q4: What types of electrophilic warheads are commonly used to target cysteine?
A4: Common electrophiles include acrylamides, α-cyanoacrylamides, alkyl halides, epoxides, aldehydes/ketones, and strained bicycloalkyl systems. Each warhead differs in reactivity, reversibility, and selectivity toward specific cysteine residues.
-
Q5: Can chemoproteomics identify cysteine sites in intact cells versus lysates?
A5: Yes. Intact cell labeling preserves the native protein conformation and cellular localization, providing more physiologically relevant data. Lysate-based labeling may allow deeper coverage but can expose normally buried cysteines.
Demo
Demo: Strategy for cysteine‑targeting covalent inhibitors screening using in‑house database based LC‑MS/MS
A LC‑MS/MS screening workflow was developed to detect potential cysteine‑targeting covalent compounds. The method incubates candidate compounds with N-acetyl-cysteine and utilizes multiple reaction monitoring (MRM) to sensitively detect cysteine adducts, facilitating the rapid identification of novel covalent binders.
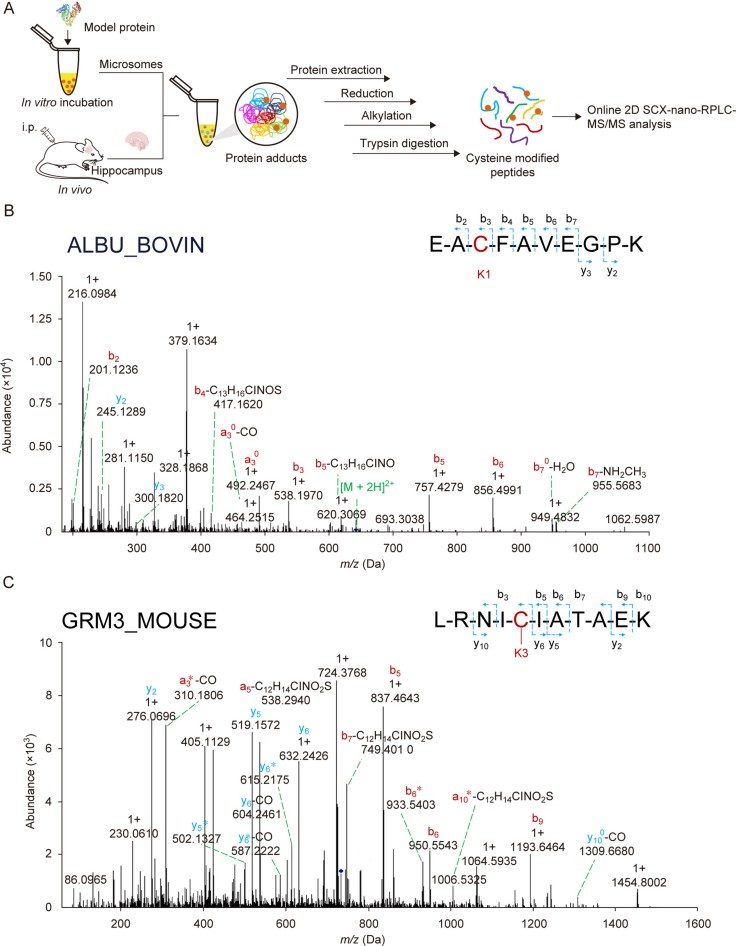
Figure 2. Proteomic strategy and identification of protein adducts formed with ketamine metabolites (Hu X, et al., 2025).
-
Case Study
Case: Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State
Background
KRAS is a frequently mutated oncogene in human cancers and has long been considered a challenging target for direct intervention. The G12C substitution introduces a nucleophilic cysteine (C12) that creates an opportunity for covalent, mutation-selective inhibitors. Prior efforts identified allosteric Switch II binders but lacked compounds with robust cellular engagement.
Purpose
Develop and characterize a cell-active, cysteine-targeted small molecule that selectively binds KRASG12C, define its mechanism of action, quantify target engagement and selectivity using MS-based assays, and test how KRAS nucleotide dynamics and upstream signaling influence inhibitor efficacy.
Methods
- The targeted LC-MS/MS peptide assay measures loss of the C12-containing tryptic peptide (quantified against isotopic standards) to report cellular covalent engagement.
- Proteome-wide cysteine profiling to assess selectivity.
- Biochemical assays and X-ray crystallography to define state specificity and binding mode.
- Cellular signaling, proliferation, and combination treatment studies to assess functional effects.
Results
- The targeted LC-MS/MS analysis demonstrated time- and dose-dependent cellular engagement, and intact-protein MS confirmed the formation of covalent adducts.
- Proteome-wide cysteine profiling showed high selectivity: KRASG12C was the most potently engaged target; very few off-targets at biologically relevant doses.
- ARS-853 treatment suppressed KRAS-effector interactions and downstream MAPK/PI3K signaling, induced cell-cycle arrest and apoptosis in KRASG12C cells, and inhibited growth in 3-D/anchorage-independent assays.
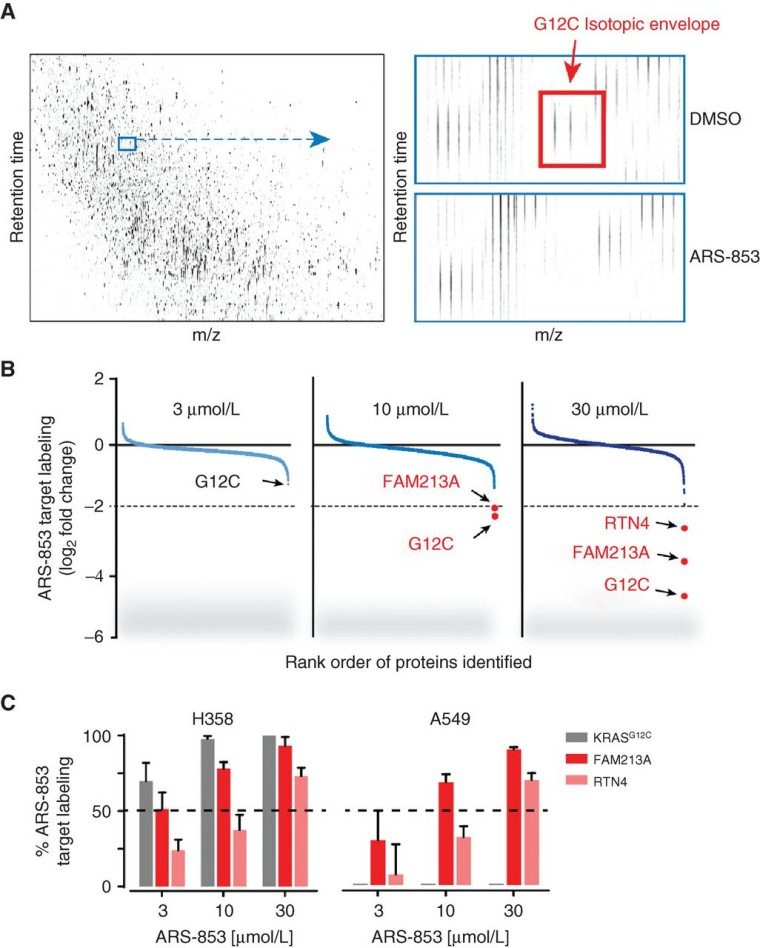
Figure 3. Proteomic cysteine profiling of ARS-853 selectivity.
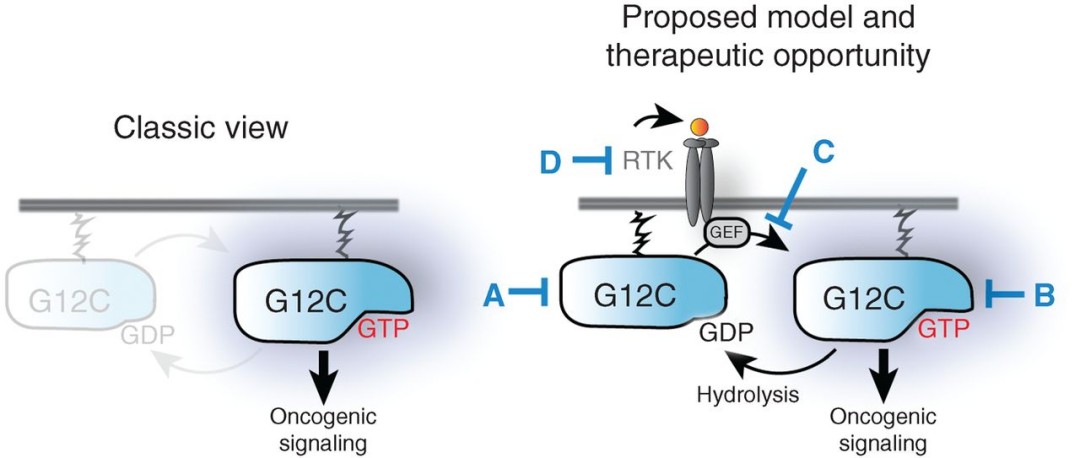
Figure 4. Schematic models of the conventional (left) and proposed model (right) for the maintenance and modulation of active mutant KRAS levels.
Conclusion
ARS-853 is a selective, cell-active covalent inhibitor of KRASG12C that preferentially targets the GDP-bound state and functionally inactivates the oncoprotein. MS-based targeted peptide quantification and proteome-wide cysteine profiling were critical for demonstrating on-target engagement, selectivity, and revealing unexpected rapid nucleotide cycling of KRASG12C These findings support targeting the inactive state of specific mutant proteins, suggesting that the upstream signaling context can significantly influence the engagement and efficacy of covalent inhibitors.
Related Services
References
- Csorba N, Abranyi-Balogh P, Keseru G M. Covalent fragment approaches targeting non-cysteine residues. Trends in Pharmacological Sciences, 2023, 44(11): 802-816.
- Patricelli M P, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer discovery, 2016, 6(3): 316-329.
- Hu X, et al. Strategy for cysteine-targeting covalent inhibitors screening using in-house database based LC-MS/MS and drug repurposing. Journal of Pharmaceutical Analysis, 2025, 15(3): 101045.


