Targeted Protein Degradation (TPD) Services
What is Targeted Protein Degradation (TPD)?
Targeted Protein Degradation (TPD) refers to approaches that leverage endogenous cellular degradation pathways to reduce the abundance of specific proteins. Instead of blocking protein activity, TPD strategies aim to remove the protein itself, enabling regulation of scaffolding proteins, multi-domain complexes, and other challenging targets.
Most TPD approaches rely on intracellular degradation systems, such as ubiquitin-dependent proteolysis or lysosome-mediated pathways. Because degradation is an event-driven process, assays must directly measure protein abundance changes over time, rather than relying solely on downstream functional readouts.
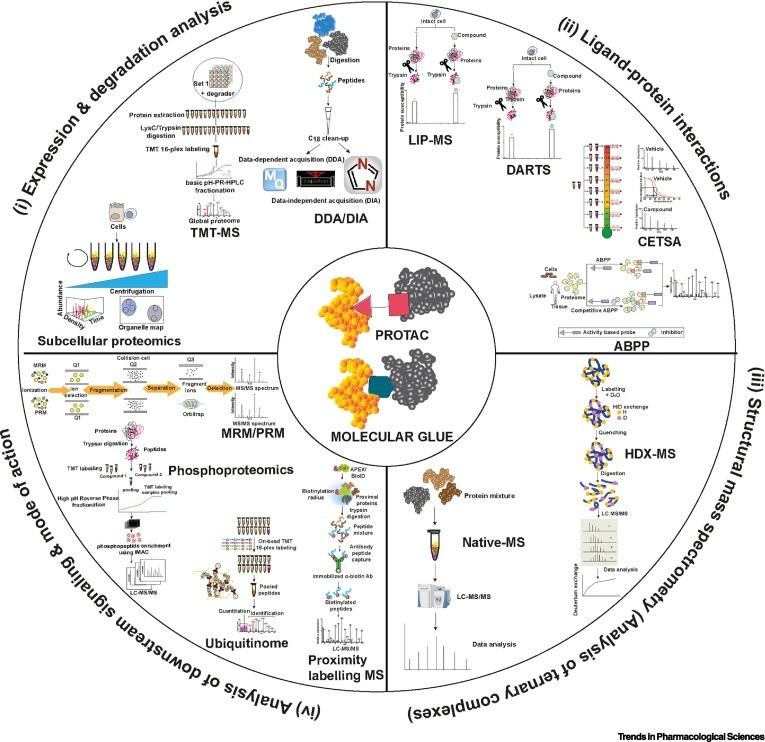
Figure 1. Comprehensive proteomics technologies for advancing targeted protein degradation (Sathe G, et al., 2023).
Targeted Protein Degradation Testing Strategies
TPD assay strategies can be broadly grouped according to project stage:
- Early screening and feasibility assessment: Primary degradation assays are commonly used to evaluate large compound sets or experimental constructs rapidly.
- Kinetic profiling and ranking of degradation performance: Beyond endpoint measurements, degradation kinetics provide deeper insight into degrader behavior.
- Proteome-wide selectivity and mechanism evaluation: Quantitative proteomics enables direct measurement of protein abundance changes across thousands of proteins simultaneously.
Advantages of Quantitative Proteomics in TPD
- Unbiased assessment of on-target and off-target degradation
- High specificity at the peptide level
- Compatibility with complex biological matrices
- Quantitative comparison across conditions and time points
Mass Spectrometry-Based Proteomics for TPD
Discovery proteomics
Best for broad off-target discovery. Works well with many samples and offers high dynamic range. Typical deliverables: protein-level fold changes, volcano plots, pathway enrichment.
Multiplexed quantitative proteomics
Isobaric labeling enables multiplexing of samples to analyze time courses or dose responses within a single run. It increases throughput and reduces run-to-run variability but requires careful correction for ratio compression.
Targeted proteomics and absolute quantitation
Once targets are defined, targeted MS yields high sensitivity and precise quantitation suitable for translational PD assays. Targeted assays are necessary for low-abundance proteins or clinical sample work.
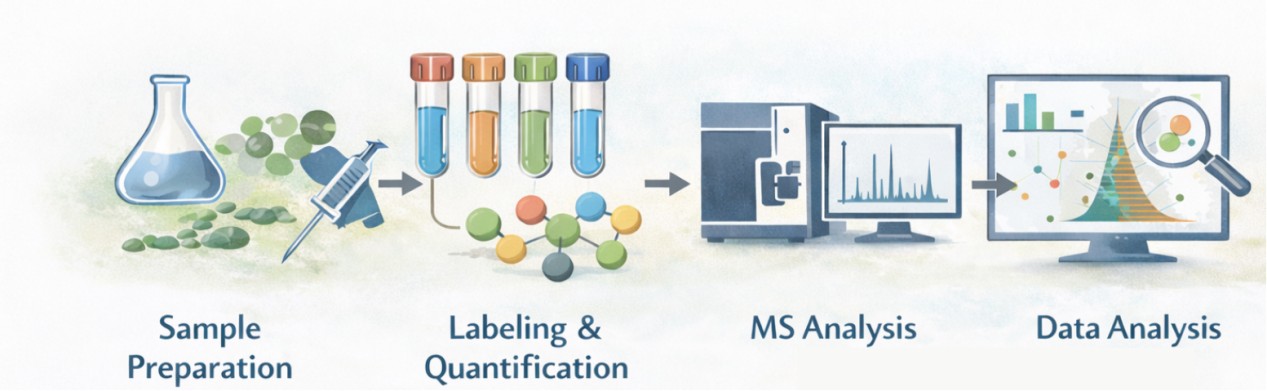
Our Mass Spectrometry Workflows for Degrader Profiling
- Sample Preparation: Cells or tissues are harvested and lysed, followed by protein extraction and enzymatic digestion.
- Labeling & Quantification: Isobaric labeling or stable isotope approaches enable multiplexed comparison.
- MS Analysis: High-resolution instruments such as Orbitrap or timsTOF provide accurate measurement of protein abundance.
- Data Analysis: Bioinformatic pipelines process quantitative results, identify significantly degraded targets, and highlight off-target effects.
Selecting the Right Service Package
Our service packages can support TPD research across stages:
- Discovery Screening: Endpoint degradation measurement, high-throughput reporter assays
- Lead Optimization: Time-resolved kinetics, MS-based profiling, off-target assessment
- Translational Studies: Multi-omic biomarker analysis, spatial and cell-specific evaluation
How Creative Proteomics Supports TPD Research
Our services provide integrated solutions for TPD studies:
- Quantitative Proteomics Services: TMT/DIA profiling, off-target analysis
- Reporter Assays: Endpoint and time-resolved degradation assessment
- Kinetic & Mechanistic Support: Ternary complex and ubiquitination evaluation
- Multi-omic Integration: Spatial and cell-specific profiling
Sample Requirements for TPD Assays
| Sample type | Typical use case | Minimum input (practical) | Recommended input | Storage / prep |
| Cultured cells (lysate) | Screening, High-Sensitivity Protein Tagging Technology, TR-Fluorescence Energy Transfer, discovery MS | 0.5–1 ×106 cells | 2–5 ×106 cells per condition | Wash, pellet, snap-freeze; store −80°C; avoid multiple freeze–thaw |
| Tissue (fresh/frozen) | Discovery MS, TMT, PRM | 2–5 mg | 10–20 mg | Snap-freeze in liquid N2; store −80°C; homogenize in appropriate buffer |
| Plasma / serum | Plasma proteomics, PK/PD, biomarker testing | 20–50 µL | 100–200 µL | Collect EDTA/heparin; separate plasma quickly; store −80°C |
Application Areas Supported by TPD Proteomics
- Measurement of target protein degradation efficiency.
- Identification of unintended protein loss.
- Comparative evaluation of degrader designs.
- Assessment of global protein homeostasis changes.
Why Choose Creative Proteomics for TPD Services
- Deep proteomics expertise: Decades of experience in proteomic analysis using mass spectrometry explicitly applied to targeted protein degradation research.
- Comprehensive quantitative proteomics services: Integrated workflows to assess degradation efficiency, kinetics, selectivity, and off-target effects with high sensitivity and reproducibility.
- Advanced mass spectrometry platforms: High-resolution, high-throughput instruments support confident protein identification and accurate quantification from complex samples.
- Method-driven experimental design: Assays are tailored to project goals, with appropriate controls and orthogonal validation to reduce experimental bias.
- Actionable and transparent data delivery: Clear, well-annotated datasets and visual summaries designed to support confident scientific decision-making.
FAQ
-
Q1: What biological systems do TPD approaches leverage?
A1: Most TPD strategies exploit the ubiquitin-proteasome system, but some emerging approaches also involve lysosome-mediated degradation pathways.
-
Q2: How does proteomic profiling distinguish on- and off-target effects?
A2: By comparing protein abundance between control and degrader-treated samples across the proteome, statistical analysis can identify proteins that are significantly degraded in a treatment-specific manner.
-
Q3: How do mass spectrometry and structural methods complement each other in TPD research?
A3: Proteomics reveals functional protein abundance changes, while structural methods like modelling or interaction studies help interpret ternary complex formation critical for degraders.
-
Q4: What are common pitfalls in TPD proteomics?
A4: Challenges include low abundance targets, incomplete proteome coverage, batch effects, and distinguishing degradation from protein synthesis inhibition.
Demo
Demo: Proteome-Wide Discovery of Degradable Proteins Using Bifunctional Molecules
large-scale TMT proteomics screen of a bifunctional library to identify proteins that can be driven to degradation; demonstrates how proteomics-first screening can expand the set of degrader-tractable proteins.
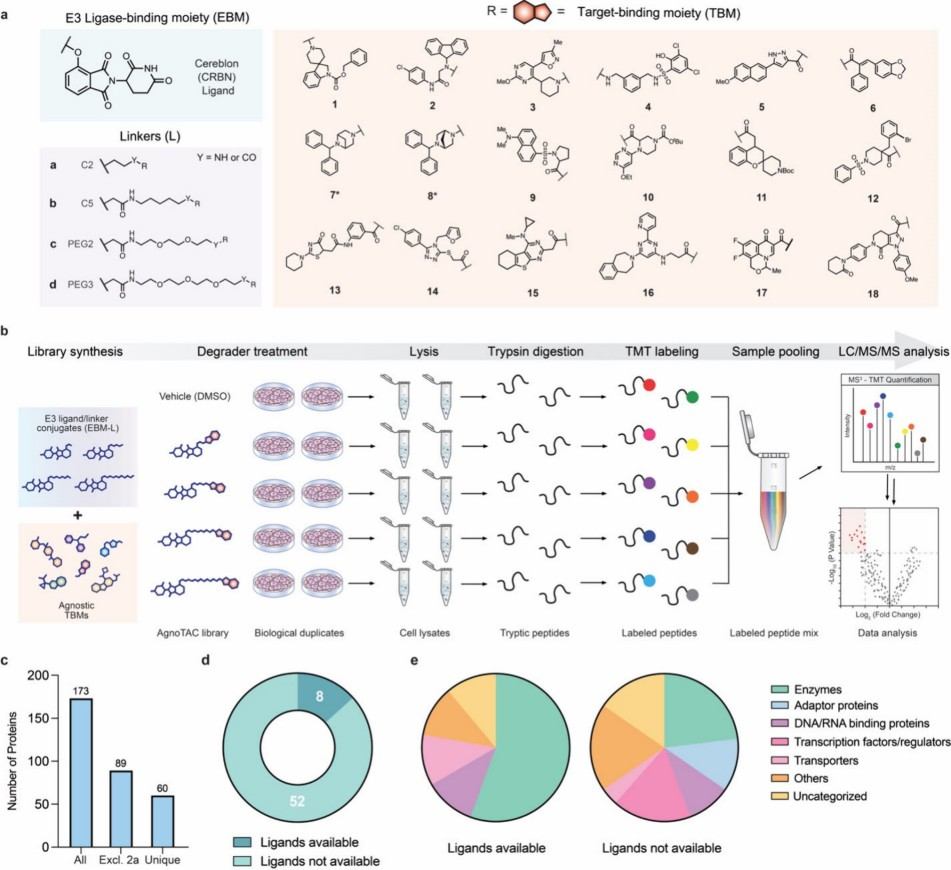
Figure 2. Chemical proteomic strategy to interrogate the degradable proteome (Forrest I, et al., 2025).
-
Case Study
Case: Degradome analysis to identify direct protein substrates of small-molecule degraders
Background
Mass spectrometry–based proteomics can measure protein abundance and ubiquitination at scale, but conventional time-course proteomics alone may conflate primary degradation with downstream effects caused by target loss. Jochem et al. develop a mass-spec workflow focused on isolating direct degradation events while excluding confounding transcriptional or translational changes.
Purpose
The study's objective was to create and validate a proteomics method that selectively quantifies proteins undergoing direct degradation after treatment with small-molecule degraders, thereby enabling confident identification of on-target substrates and distinguishing them from secondary proteome responses. The method aims to be broadly applicable across degrader modalities and to produce high-confidence substrate lists for downstream mechanistic studies.
Methods
- Metabolic pulse labelling: Cells are pulse-labelled with an azidohomoalanine (AHA) or by SILAC-based approaches to mark newly synthesized proteins prior to/after degrader treatment.
- Click chemistry enrichment: AHA-labelled proteins (or isotopically labelled channels) are selectively enriched via click chemistry, isolating pools that reflect nascent synthesis and allowing subtraction of synthesis-driven changes.
- Quantitative LC–MS/MS: Enriched samples are analyzed by high-resolution LC–MS/MS with isobaric or metabolic labelling quantitation to compare treated vs control conditions at defined time points.
- Orthogonal validation: The approach is benchmarked against known degraders and validated using proteasome inhibitors and genetic perturbations when appropriate.
Results
- Method performance: DegMS robustly identifies directly degraded substrates and reduces false positives that would arise from transcriptional/translational down-regulation.
- Application to known degraders: When applied to established degrader molecules, the method recapitulated known primary substrates and revealed additional, previously unrecognized direct targets in the same datasets.
- Case studies / mechanistic insight: The authors demonstrate that combining DegMS with proteasome inhibition and orthogonal assays helps confirm that identified hits are processed via the expected degradation pathway.
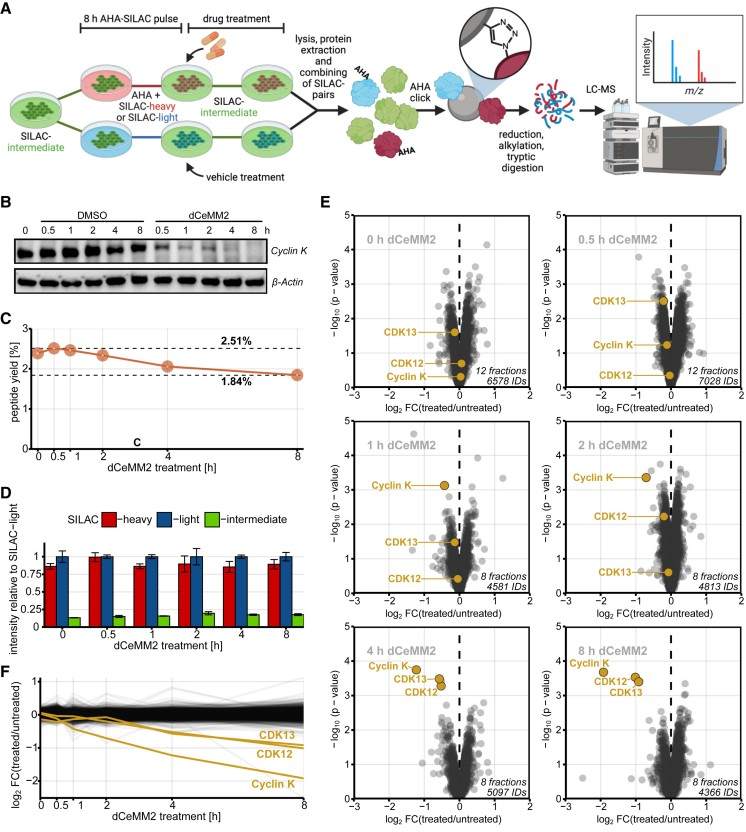
Figure 3. Degradome proteomics by DegMS.
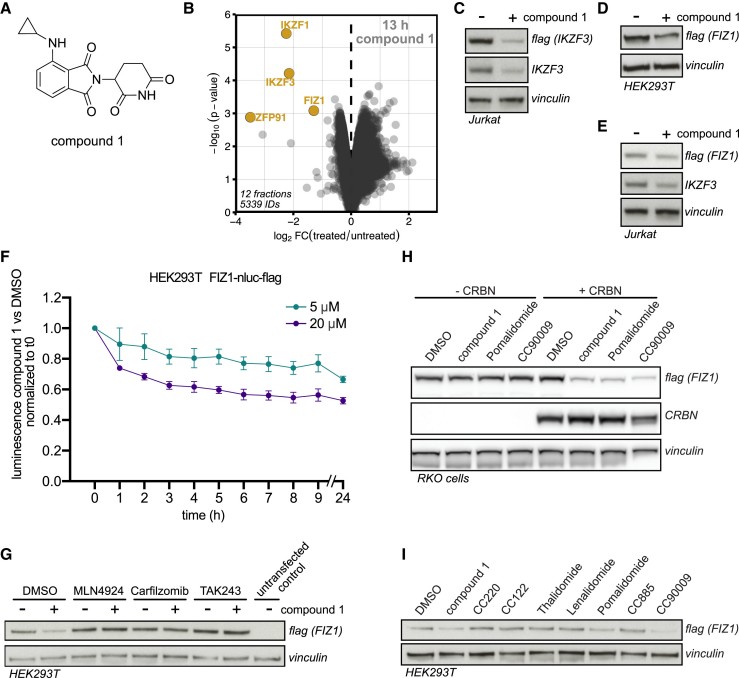
Figure 4. Benchmarking of degradome proteomics by DegMS.
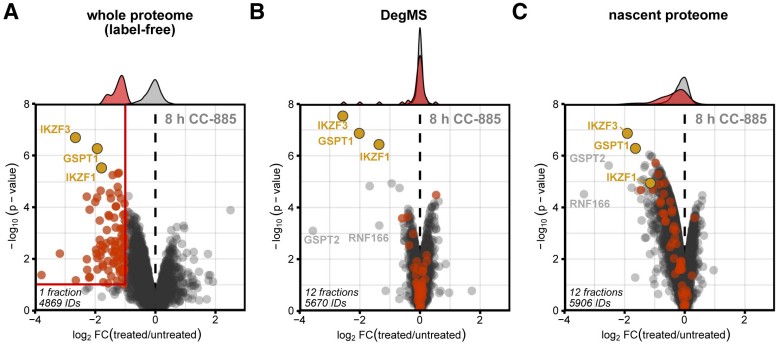
Figure 5. Proteome response to CC-885 treatment.
Conclusion
DegMS is a scalable, MS-based strategy that selectively captures proteins undergoing direct degradation in response to small-molecule degraders while excluding confounding synthesis changes. This yields higher-confidence substrate lists and helps distinguish primary molecular targets from downstream proteome effects. The paper demonstrates the method's applicability across different degrader chemistries and provides deposited datasets for community reuse, making it a practical template for labs and CROs that need mechanistic substrate identification in degrader projects.
Related Services
References
- Sathe G, Sapkota G P. Proteomic approaches advancing targeted protein degradation. Trends in pharmacological sciences, 2023, 44(11): 786-801.
- Forrest I, et al. Proteome-Wide Discovery of Degradable Proteins Using Bifunctional Molecules. ACS Central Science, 2025, 11(11): 2240-2256.
- Jochem M, et al. Degradome analysis to identify direct protein substrates of small-molecule degraders. Cell chemical biology, 2025, 32(1): 192-200. e6.