Tumor Neoepitope Identification Services
Why Does Neoepitope Matter in Cancer Immunotherapy?
Neoepitopes are short, tumor-specific peptides produced when cancer cells carry unique mutations or make/process proteins abnormally. Because they do not occur in normal tissues, the immune system recognizes them as foreign, and they reliably provoke immune responses.
Accurate identification of true neoepitopes is crucial for research, as it determines which targets are suitable for vaccine design, engineered T-cell studies, or as markers of immune activity. Computational prediction alone yields many false positives; experimental proof that a peptide is actually processed and presented on tumor cells is needed to focus on the candidates that matter.
Why Choose MS-Based Immunopeptidomics
There are two major techniques for identifying neoantigen epitopes: the immunogenomic or immunopeptidomic method. Mass spectrometry (MS)-based immunopeptidomics enables the direct identification of MHC-presented peptides from tumor samples. By isolating MHC complexes and sequencing their bound peptides using high-resolution LC-MS/MS, this approach captures neoepitopes that are naturally processed and displayed on the tumor cell surface. Compared with prediction-only pipelines, immunopeptidomics offers several advantages:
- Direct confirmation of antigen presentation.
- Reduced false-positive neoepitope candidates.
- Detection of low-abundance and non-canonical peptides.
- Higher translational confidence for downstream therapeutic development.
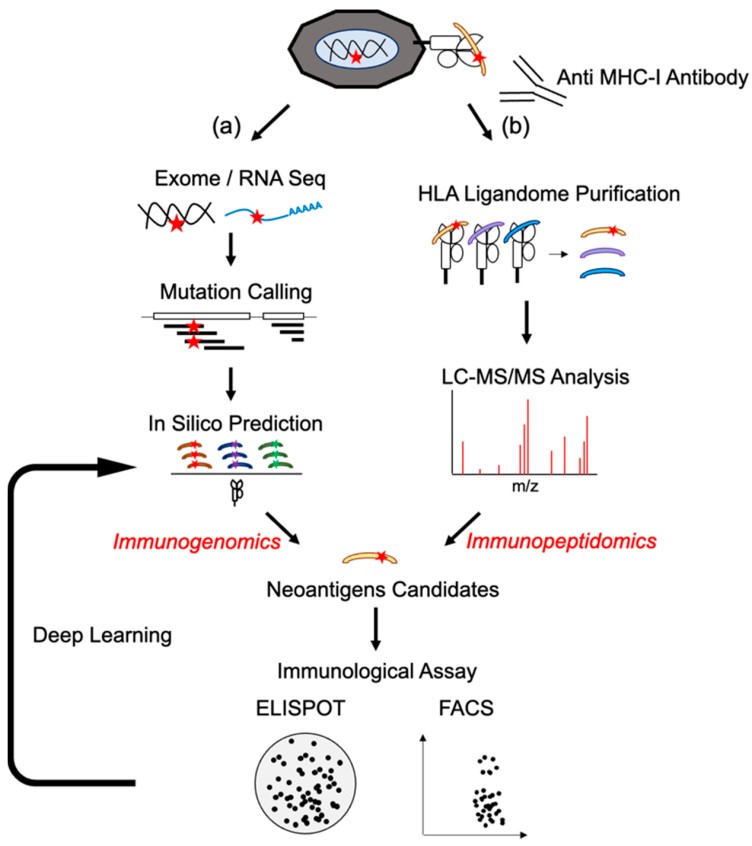
Figure 1. Neoantigen identification by immunogenomic or immunopeptidomic method (Okada M, et al., 2022).
Integrated Multi-Omics Strategy for High-Confidence Neoepitope Discovery
High-confidence neoepitope identification requires more than peptide detection alone. At Creative Proteomics, we integrate proteomics, genomics, and transcriptomics to ensure that each reported neoepitope is biologically real, tumor-specific, and relevant for downstream immunotherapy development.
MS–based immunopeptidomics directly identifies peptides that are naturally displayed on tumor cell surfaces by MHC molecules. This confirms that a neoepitope is actually presented to the immune system.
Genomic analysis (such as whole-exome or whole-genome sequencing) links each identified peptide to its underlying DNA mutation. This step verifies that the neoepitope originates from a tumor-specific genetic change and is absent from normal tissues.
Transcriptomic data (RNA sequencing) adds a third layer of confidence by confirming that the mutated gene is actively expressed in the tumor. Neoepitopes derived from genes that are not expressed are unlikely to be presented at meaningful levels and are therefore deprioritized.
Bioinformatics Pipeline: From Raw Spectra to Prioritized Neoepitopes
Key Analytical Steps:
- Somatic mutation calling and annotation
- Customized proteogenomic database construction
- Peptide spectrum matching with stringent FDR control
- HLA typing and MHC binding affinity prediction
- Neoepitope prioritization based on presentation confidence, expression, and immunogenic potential
Advantages of a Proteomics-Driven Neoepitope Strategy
Compared with sequence-based prediction alone, proteomics-driven neoepitope identification offers:
- Experimental confirmation of peptide presentation
- Reduced false-positive candidate lists
- Access to non-canonical antigen sources
- Improved biological relevance for downstream research
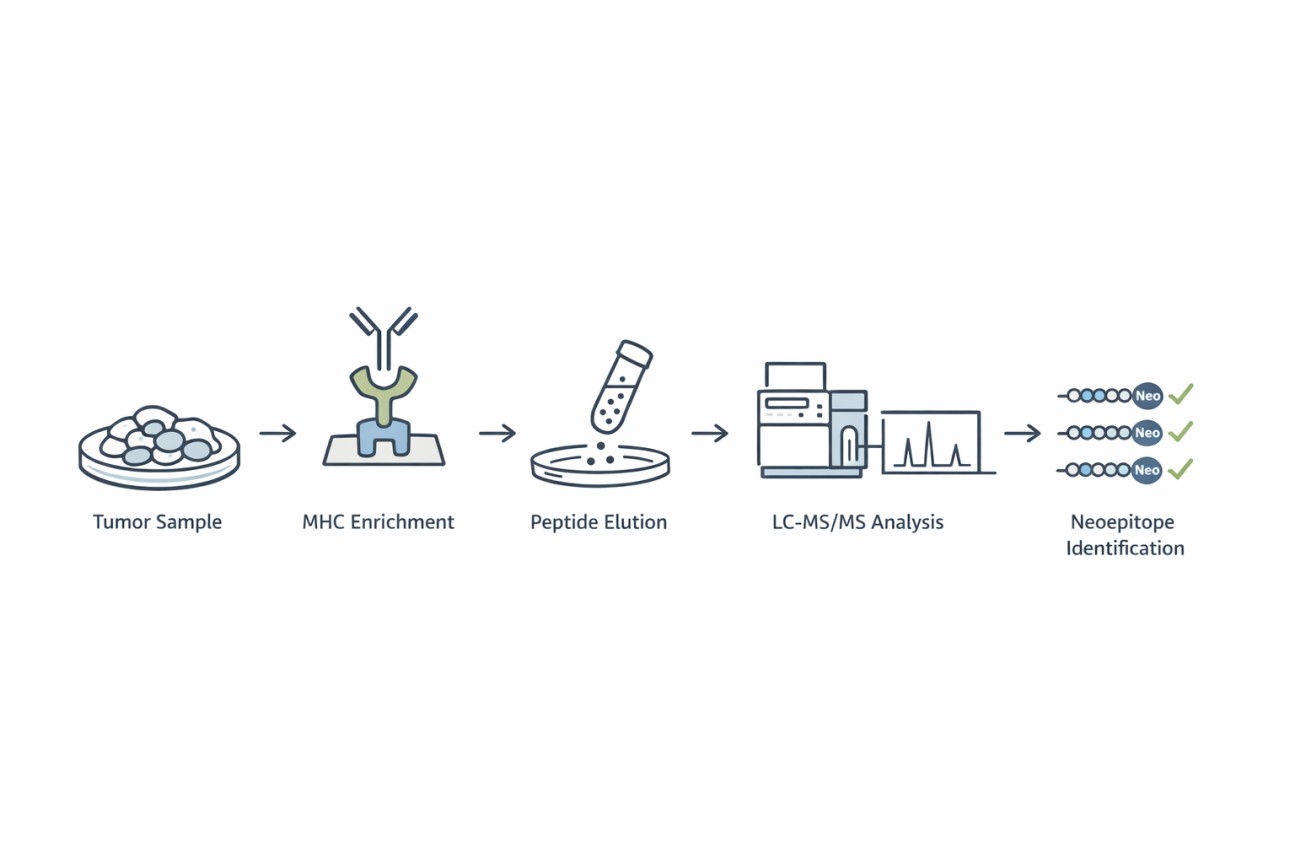
Creative Proteomics' Neoepitope Identification Service Workflow
- MHC Enrichment: High-affinity monoclonal antibodies are used to isolate MHC class I and/or class II complexes from tumor samples.
- Peptide Elution: MHC-bound peptides are gently eluted without disrupting peptide integrity.
- LC-MS/MS Analysis: High-resolution mass spectrometry identifies peptide sequences with high sensitivity and accuracy.
- Bioinformatic Annotation: Identified peptides are matched against customized proteogenomic databases to distinguish neoepitopes from self-peptides.
Deliverables: What You Receive
Clients receive a comprehensive and actionable dataset, including:
- Identified MHC-bound peptide lists with MS evidence
- Annotated variant files (VCF/TSV)
- HLA typing results
- Prioritized neoepitope candidates
- Summary reports with interpretation guidance
Validation Strategies and Downstream Support
To further increase confidence, Creative Proteomics offers optional validation strategies:
- Targeted MS validation (PRM/SRM)
- Synthetic peptide confirmation
- Support for T-cell reactivity assays
Applications in Cancer and Immunology Research
- Studying antigen processing and presentation mechanisms
- Investigating tumor immune evasion strategies
- Comparing neoepitope landscapes across tumor subtypes
- Supporting exploratory immune profiling studies
Sample Requirements for Tumor Neoepitope Identification
| Sample Type | Recommended Input | Key Requirements |
| Tumor Tissue (Fresh or Frozen) | ≥ 100–300 mg | High cellularity; minimal necrosis; rapid freezing after collection. |
| Tumor Cell Lines | ≥ 1 × 10⁷ cells | High viability (>90%); standardized culture conditions. |
| Peripheral Blood Mononuclear Cells (PBMCs) | ≥ 5 × 10⁷ cells | Fresh or cryopreserved; minimal freeze–thaw cycles. |
| Matched Normal Tissue (Optional) | ≥ 50–100 mg | Same processing workflow as tumor tissue. |
Why Choose Creative Proteomics for Neoepitope Identification
- Over years of proteomics and mass spectrometry expertise.
- State-of-the-art Orbitrap and timsTOF platforms.
- Integrated proteogenomic and multi-omics workflows.
- High-confidence, experimentally validated neoepitope discovery.
- Dedicated scientific support from project design to data interpretation.
FAQ
-
Q1: Why integrate genomics/transcriptomics (WES/WGS, RNA-seq) with immunopeptidomics (i.e., proteogenomics)?
A1: Integrating sequencing data with MS enables the construction of custom protein databases that include patient- or sample-specific variant sequences, splice isoforms, and non-canonical translation products. This proteogenomic approach increases the chance of correctly matching spectra to mutation-derived peptides and expands discovery beyond reference proteomes.
-
Q2: What are the principal limitations and common sources of false negatives/positives?
A2: Common limitations are (a) low abundance of presented neoepitopes below MS detection limits, (b) incomplete or incorrect variant calling or database generation, (c) peptide losses during enrichment or sample handling, and (d) expanded search spaces when looking for non-canonical peptides, which can inflate false positives unless FDR is carefully controlled.
-
Q3: How to distinguish true neoepitopes from background self-peptides?
A3: Experimental strategies include: (1) using matched normal controls to subtract peptides presented in healthy cells; (2) motif analysis and HLA allele specificity checks; (3) comparison to public immunopeptidome databases; and (4) statistical filtering based on spectral counts and reproducibility across replicates.
Demo
Demo: Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry
This study mapped the peptides presented on HLA molecules in human melanoma tissues using high-resolution mass spectrometry. Tens of thousands of peptides were analyzed, and 11 mutation-derived neoepitopes were identified from the tumors. Some of these peptides triggered T-cell responses, showing that mass spectrometry can detect naturally presented neoepitopes without relying only on predictions.
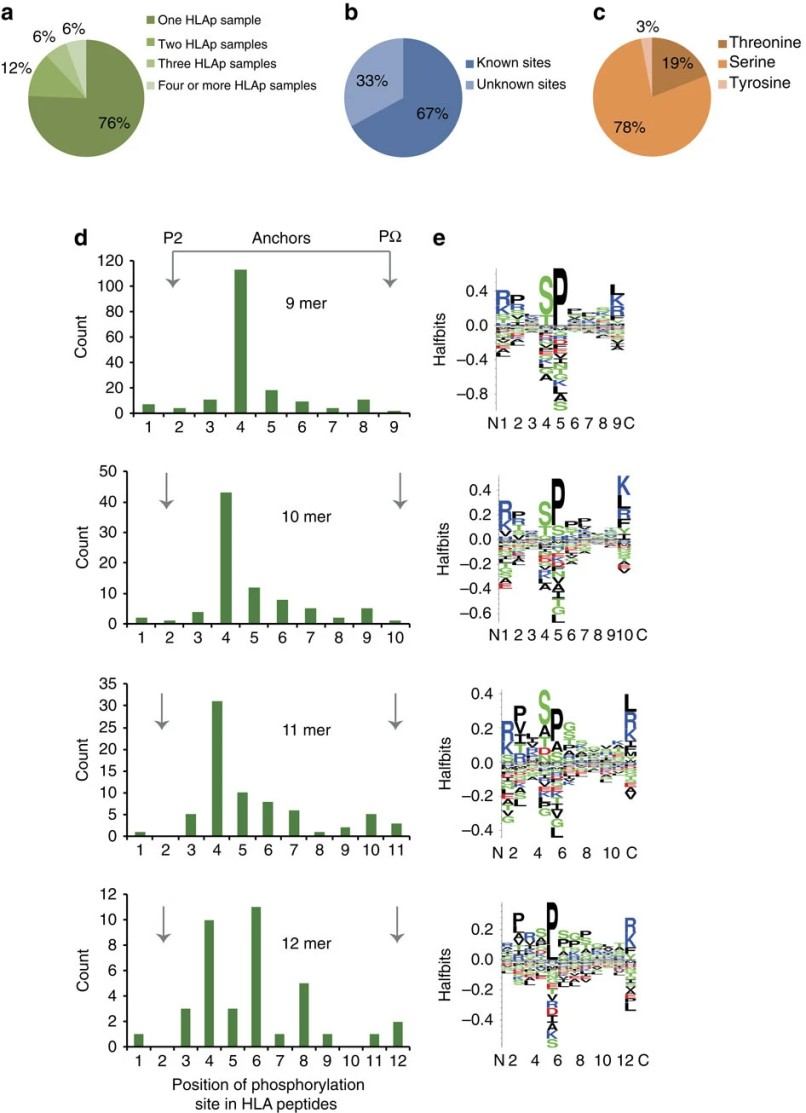
Figure 2. Characterization of phosphorylation on eluted HLA peptides (Minegishi Y, et al., 2016).
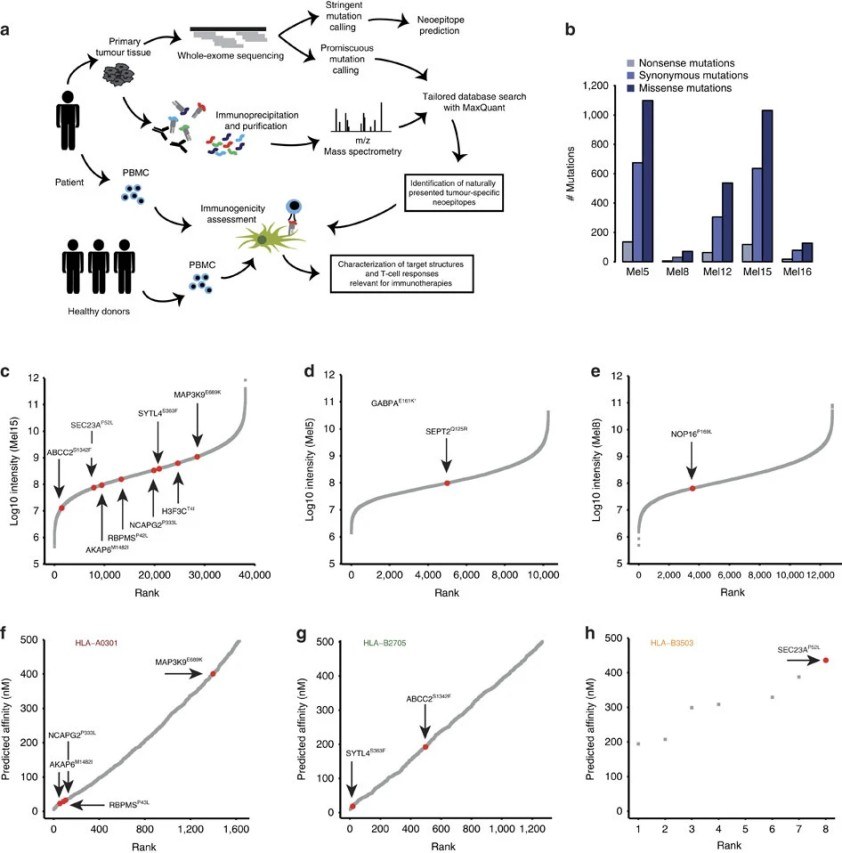
Figure 3. Identification of mutated peptide ligands by matching exome sequencing and mass spectrometry immunopeptidomics (Minegishi Y, et al., 2016).
-
Case Study
Case: Differential ion mobility mass spectrometry in immunopeptidomics identifies neoantigens carrying colorectal cancer driver mutations.
Abstract
This study developed a highly sensitive immunopeptidomics workflow to detect neoepitopes in primary human colorectal cancer tissues directly. By combining advanced ion mobility separation with mass spectrometry, the method overcomes challenges in identifying low-abundance mutation-derived peptides, enabling more reliable neoepitope discovery from real tumor samples.
Methods
- Sample Collection: Tumor and adjacent normal tissues were collected from patients with colorectal cancer (n=17).
- Proteogenomic Analysis: Customized peptide databases were constructed using both normal human proteome and patient‑specific somatic mutation information.
- Peptide Identification: Tandem mass spectra were matched to the tailored database to identify MHC‑presented peptides, including mutation‑derived neoantigens.
- Validation: Identified neoepitopes were further confirmed using synthetic peptide standards with targeted acquisition modes (e.g., PRM).
Results
- Comprehensive Immunopeptidome: Across 17 paired colorectal tumor and normal samples, an average of ~4,921 immunopeptides were identified per patient.
- Neoepitope Detection: Among the thousands of immunopeptides, two specific neoantigen peptides arising from clinically relevant driver mutations—KRAS‑G12V and CPPED1‑R228Q—were detected and confirmed by spectral matching with synthetic standards.
- Tumor Specificity: The direct identification of mutant peptides from tumor tissues highlighted differential antigen processing compared with corresponding normal tissues.
- Workflow Feasibility: FAIMS‑assisted differential ion mobility MS enabled sensitive discovery of neoantigens even from limited tissue material, demonstrating its utility in high‑confidence immunopeptidomics studies
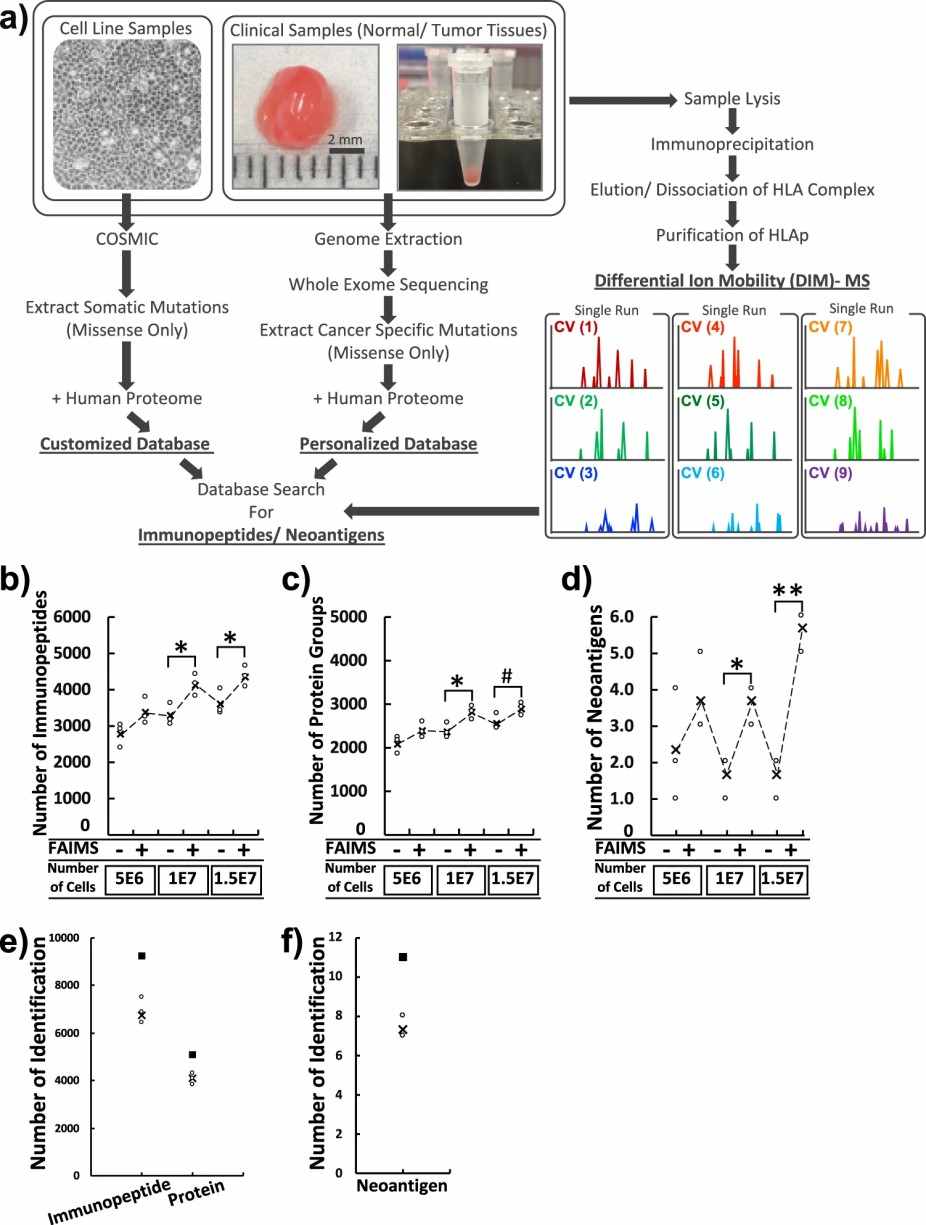
Figure 4. A schematic workflow and validation of global-immunopeptidomics analysis.
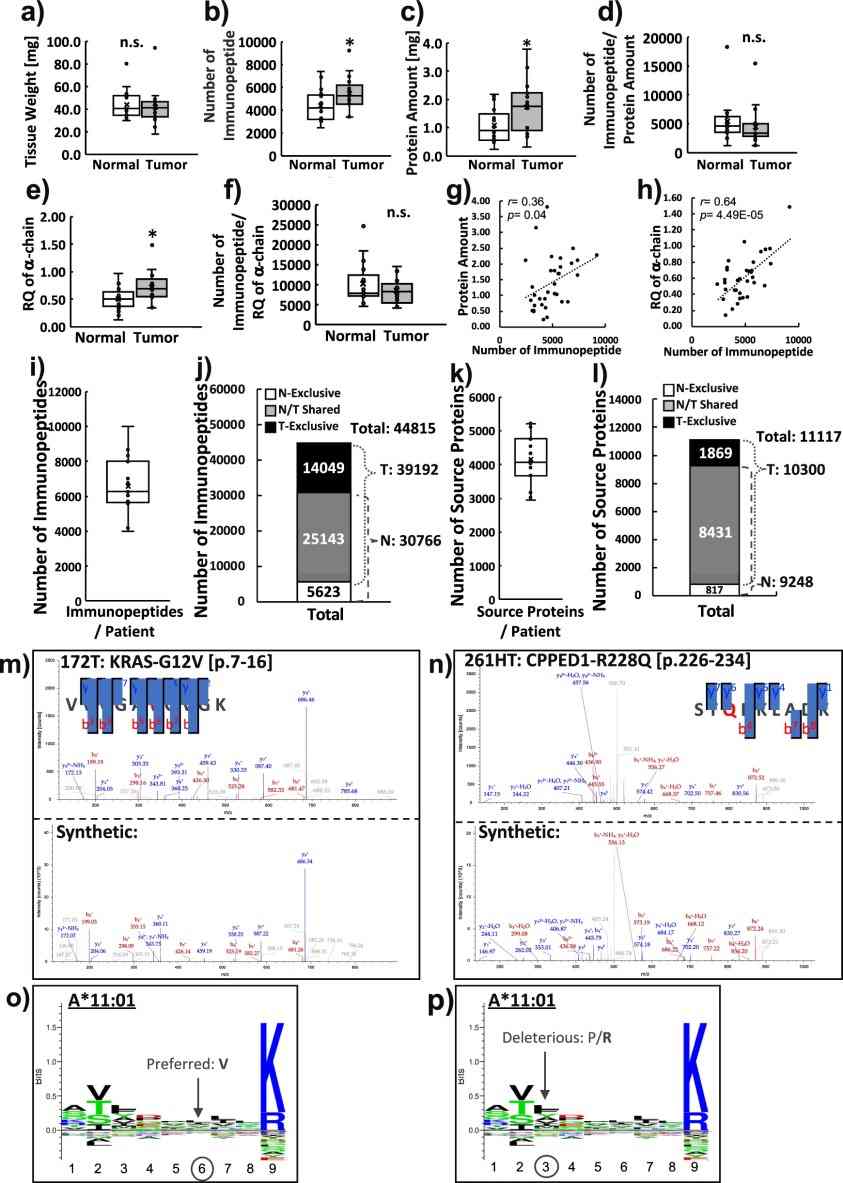
Figure 5. Personalized immunopeptidomics analyses from 17 colorectal cancer patients.
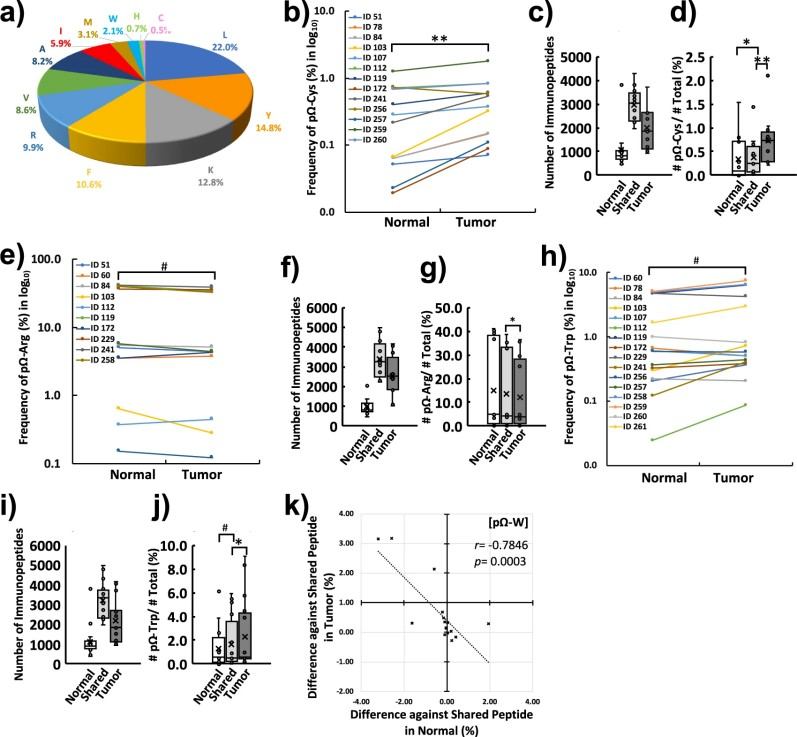
Figure 6. The CRC immunopeptidome, neoantigens directly identified from tissues and the cancer-specific profiling of HLAp by global immunopeptidomics analysis.
Conclusion
This study provides evidence that immunopeptidomics can successfully identify mutation‑derived neoepitopes directly from human tumor tissues, including those associated with common driver mutations in colorectal cancer. The work demonstrates that advanced MS workflows can overcome challenges related to low abundance and complexity of native immunopeptidomes, supporting deeper exploration of naturally presented neoepitopes in cancer research.
Related Services
References
- Okada M, Shimizu K, Fujii S. Identification of neoantigens in cancer cells as targets for immunotherapy. International Journal of Molecular Sciences, 2022, 23(5): 2594.
- Minegishi Y, et al. Differential ion mobility mass spectrometry in immunopeptidomics identifies neoantigens carrying colorectal cancer driver mutations. Communications Biology, 2022, 5(1): 831.
- Bassani-Sternberg et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nature communications, 2016, 7(1): 13404.