PDX/CDX Models Proteomic Characterization
How to Advance Translational Research with PDX/CDX Proteomics
Patient-derived xenograft (PDX) and cell line-derived xenograft (CDX) animal models are foundational tools in modern oncology and drug discovery research. They enable investigators to evaluate tumor biology, therapeutic response, and resistance mechanisms within physiologically relevant in vivo systems.
While genomic and transcriptomic profiling provide valuable molecular context, proteomic characterization captures functional biology directly, reflecting pathway activation, post-translational regulation, and drug-target engagement. Quantitative proteomics therefore plays a central role in translational studies, drug discovery, and preclinical assessment.
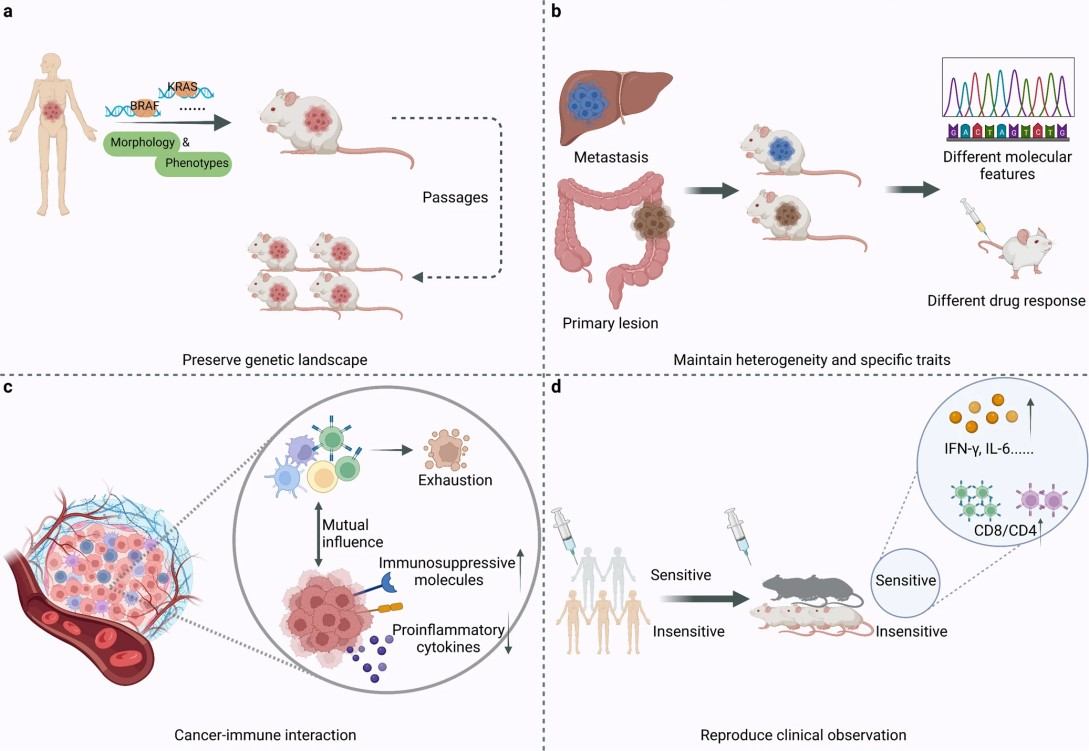
Figure 1. The advantages of PDX over other models (Liu Y, et al., 2023).
Creative Proteomics offers comprehensive proteomic characterization services for PDX and CDX animal models, combining advanced mass spectrometry, robust quantitative strategies, and expert bioinformatics to support clinical research pipelines and R&D programs.
What Is the Difference Between CDX and PDX Models?
| Feature | PDX Models | CDX Models |
| Source of Tumor Material | Established by directly implanting tumor tissue from a patient into immunodeficient or humanized mice. | Derived from commercially available or in-house cultured tumor cell lines. |
| Biological Fidelity | High fidelity to the original human tumor. Maintains histopathology, genetic heterogeneity, tumor microenvironment characteristics, and signaling networks. | Moderate biological fidelity. Tumor cell lines are often more homogeneous and may have adapted to in vitro culture conditions. |
| Experimental Throughput | Lower throughput due to limited availability of patient tissue and slower tumor engraftment. | High throughput. Cell lines are readily expandable and allow rapid establishment of multiple parallel experiments. |
| Tumor Heterogeneity | Captures intra- and inter-patient heterogeneity, enabling study of diverse molecular subtypes and resistance mechanisms. | Less heterogeneous. CDX models generally represent a single clone, providing more controlled and reproducible experimental conditions. |
| Applications | Optimal for translational studies, biomarker identification, and validation of therapeutic strategies in clinically relevant models. | Well-suited for drug screening, target validation, pharmacokinetics/pharmacodynamics (PK/PD) studies, and rapid evaluation of multiple compounds or treatment regimens. |
| Time and Cost Considerations | Longer engraftment times and higher costs due to tissue acquisition and animal maintenance. | Faster to establish and lower cost, making CDX models practical for larger-scale studies and early-stage preclinical evaluation. |
| Integration with Proteomics | Provides highly relevant protein expression and pathway profiling reflecting patient-specific tumor biology, supporting translational and preclinical assessment. | Supports reproducible proteomic profiling across multiple experiments, enabling comparative studies and validation of targets or pathways identified in PDX models. |
Quantitative Proteomics Strategies for PDX/CDX Studies
Quantitative proteomics is a core experimental approach for characterizing PDX and CDX animal models at the protein level. It enables systematic measurement of protein abundance changes within tumor tissues, providing a direct view of molecular processes operating in vivo. In PDX/CDX studies, quantitative proteomics is commonly used to investigate treatment-induced effects, compare tumor subtypes, and explore biological differences across models under controlled experimental conditions.
Label-Free Quantitative Proteomics
Label-free quantification relies on repeated measurement of peptide signal intensities or spectral counts across multiple mass spectrometry runs. In PDX/CDX studies, it is frequently employed when analyzing large numbers of xenograft tumors or when sample availability varies across different models.
Sample preparation, chromatographic conditions, and instrument parameters are tightly controlled to ensure that differences in signal reflect biological variation rather than technical noise. Quantification is typically performed using intensity-based algorithms that align peptide features across runs and calculate relative protein abundance across experimental groups.
Label-free workflows are well suited for discovery-oriented studies:
- Mapping global protein expression patterns.
- Identifying pathways altered by compound exposure.
- Comparing baseline proteomic profiles across different PDX or CDX models.
Label-Based Quantitative Proteomics
Label-based strategies enable the simultaneous analysis of multiple samples in a single mass spectrometry run. Each sample is assigned a chemical tag, enabling precise relative comparison across experimental groups. In PDX/CDX experiments, label-based strategies are often used when an accurate, direct comparison between defined conditions is required. Tumor samples are processed in parallel, labeled individually, pooled, and analyzed together to reduce experimental variability.
This strategy is particularly useful for:
- Direct comparison across treatment arms is required.
- Subtle protein changes need to be measured with high confidence.
- Batch-to-batch variability must be minimized.
Creative Proteomics' Label-based quantitative proteomics services:
- TMT-based Quantitative Proteomics Analysis
- iTRAQ-based Quantitative Proteomics Analysis
- SILAC-based Quantitative Proteomics Analysis
DIA-Based Proteomics for PDX/CDX Profiling
Data-independent acquisition (DIA) has become a core strategy for proteomic analysis of PDX and CDX animal models because it delivers consistent, reproducible, and comprehensive protein measurements across complex in vivo samples. Unlike traditional data-dependent methods that selectively analyze only the most abundant peptides, DIA systematically fragments all detectable peptides within defined mass ranges, ensuring that biologically relevant proteins are not missed due to sampling bias.
Why DIA is Well-Suited for PDX/CDX Studies
PDX and CDX tumor tissues are inherently heterogeneous and often available in limited amounts. DIA addresses these challenges by providing:
- High data completeness, allowing the same proteins to be quantified across all models and treatment groups.
- Excellent run-to-run reproducibility, which is essential for comparing multiple animal cohorts.
- Stable quantification of low-abundance signaling proteins, including kinases and pathway regulators that drive tumor behavior.
Improved Throughput Without Sacrificing Biological Depth
Modern DIA workflows enable high sample throughput while maintaining deep proteome coverage. This allows large PDX or CDX panels to be analyzed within practical timelines, supporting:
- Head-to-head comparison of multiple tumor models
- Longitudinal studies tracking molecular changes over time
- Efficient screening of treatment and control groups
Reliable Quantification for Translational Research
Because DIA generates highly consistent quantitative data, it is well suited for downstream biological interpretation. Protein-level changes can be confidently linked to altered cellular pathways, enabling clearer insights into mechanism of action, pathway activation, and potential biomarker candidates. When combined with genomic or transcriptomic data, DIA-based proteomics strengthens the functional understanding of PDX and CDX models in translational research settings.
Applications of PDX/CDX Proteomic Characterization
Biomarker Discovery for Translational Research
Proteomic analysis of PDX and CDX models enables the systematic identification of proteins whose abundance or activity changes in response to disease progression or therapeutic intervention.
Mechanism-of-Action and Target Validation
Proteomic characterization provides direct evidence of how a test compound influences cellular pathways within PDX and CDX tumors. Measuring changes in protein abundance and signaling activity allows researchers to confirm whether a compound is affecting its intended biological target.
Model Selection and Biological Relevance Assessment
Not all PDX or CDX models are equally representative of human disease. Proteomic profiling allows objective comparison of models based on their functional molecular features rather than histology alone.
Therapy Response and Resistance Analysis
Quantitative proteomics enables detailed comparison of responding and non-responding PDX/CDX tumors following treatment. These analyses can uncover protein changes associated with reduced sensitivity, adaptive signaling, or survival mechanisms.
Sample Preparation and Our Workflow
- Tissue Collection and Preservation: Tumor fragments are collected from xenografted mice and preserved to retain native protein profiles.
- Protein Extraction and Digestion: High-efficiency lysis and enzymatic digestion generate peptides suitable for high-resolution mass spectrometry.
- Quantitative Approaches: Both label-based (TMT, iTRAQ) and label-free methods are applied, depending on study design.
- Data Acquisition and Analysis: DIA or DDA acquisition modes on Orbitrap or timsTOF platforms provide comprehensive proteome coverage.
Our Validated PDX and CDX Models
| Validated PDX Models | |||
| Broad solid tumor | Colorectal, Lung (NSCLC, SCLC), Breast (including TNBC), Pancreatic, Ovarian, Gastric, Liver (HCC), Head & Neck, Esophageal, Bladder, Prostate, Kidney. | ||
| Rare/High-need indications | Sarcoma, Mesothelioma, Merkel cell carcinoma, Neuroblastoma, GIST, Pediatric tumor models. | ||
| Hematologic PDXs | AML, ALL, lymphoma subtypes (low-passage models preserving cellular hierarchy). | ||
| Tumor microenvironment–aware models | Orthotopic PDXs and disseminated models for metastasis and TME studies. | ||
| Validated CDX Models (Representative Cell Lines & Features) | |||
| Breast | MCF-7, MDA-MB-231, BT-474, SK-BR-3. | ||
| Colorectal | HCT116, HT-29, SW480, LoVo, DLD-1. | ||
| Lung | A549, H1975, H1299, H460 (NSCLC panels); DMS series for SCLC. | ||
| Prostate & GU | LNCaP, PC-3, DU145, 22Rv1. | ||
| Pancreatic | PANC-1, MIA PaCa-2, BxPC-3. | ||
| Ovarian & Gynecologic | OVCAR-3, OVCAR-8, SK-OV-3, TOV-21G. | ||
| Hematologic cell lines | HL-60, MV4-11, MV4-11, MOLM-13, K562 (leukemia/lymphoma panels). | ||
| Liver & GI | HepG2, Hep3B, SW480. | ||
| Melanoma & Skin | A375, SK-MEL-28, LOX. | ||
How Creative Proteomics Supports PDX/CDX Proteomic Analysis
- Tailored Study Design: Guidance on selecting relevant PDX/CDX models and experimental planning.
- Advanced Mass Spectrometry: Orbitrap Astral and timsTOF HT for deep proteome coverage.
- Quantitative Proteomics Workflows: Label-free and label-based approaches with DIA acquisition.
- Integrated Bioinformatics: Pathway analysis, biomarker identification, and model comparison.
FAQ
-
Q1: How do proteomic profiles in PDX models compare to their original human tumors?
A1: Proteomic studies indicate that many protein expression patterns in PDXs broadly reflect those of the original tumors, but certain pathways and post‑translational modifications may diverge due to host influences and serial passaging.
-
Q2: What types of sample preparation are optimal for xenograft proteomics?
A2: Optimized protocols include careful tumor tissue homogenization, reduction of host stromal contamination, and peptide fractionation to ensure deep, reproducible proteome coverage across samples.
-
Q3: Do proteomic differences between PDX and CDX models impact translational interpretation?
A3: Yes. PDX models better preserve tumor heterogeneity, while CDX models offer consistency; proteomic profiling reveals these differences and guides appropriate model usage depending on research goals.
Demo
Demo: PDX models reflect the proteome landscape of pediatric acute lymphoblastic leukemia but diverge in select pathways
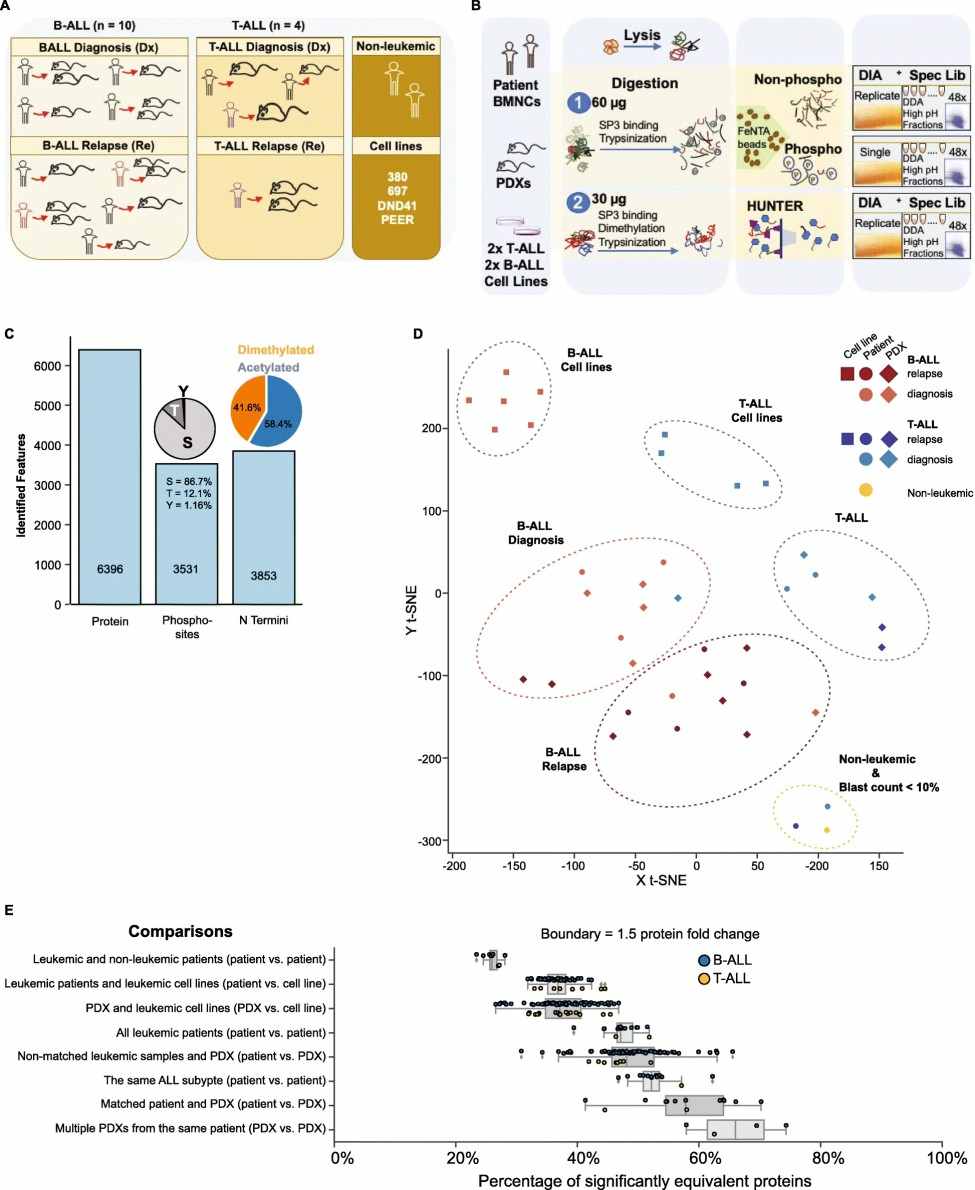
Figure 2. Proteomics stratifies pediatric ALL subtypes.
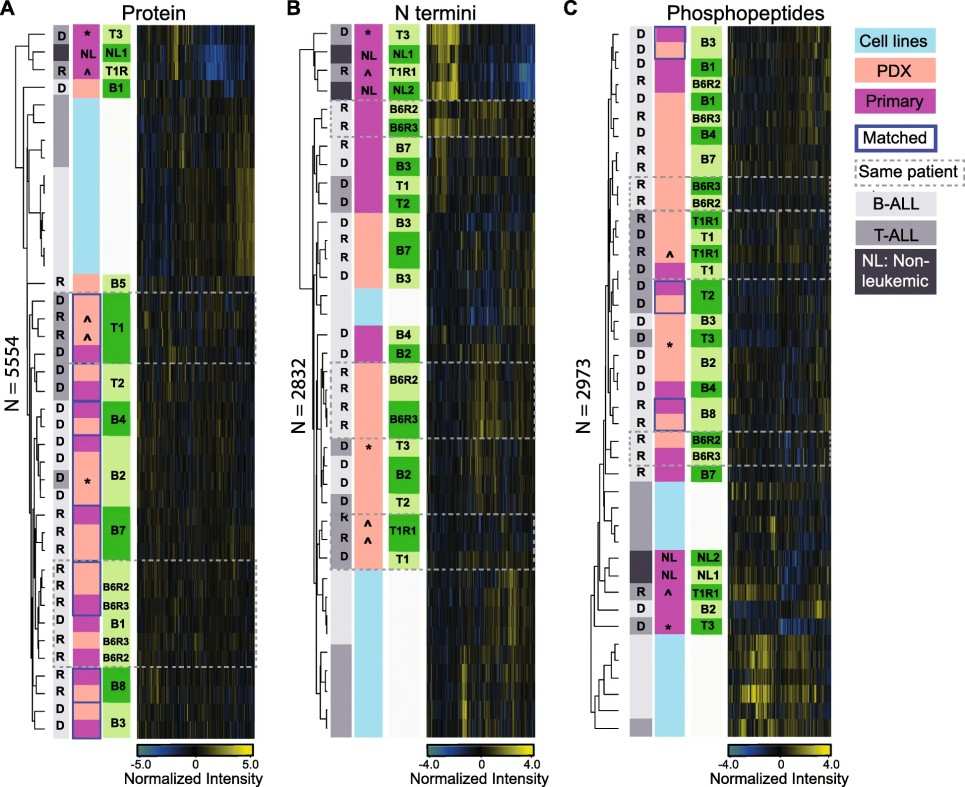
Figure 3. Pediatric ALL proteome landscape in patients, PDXs and leukemic cell lines.
-
Case Study
Case: Proteogenomic integration reveals therapeutic targets in breast cancer xenografts
Abstract
This study combined quantitative proteomics with genomic and transcriptomic data across 24 breast cancer PDX models to map protein-level differences between intrinsic cancer subtypes. By measuring proteins directly, the analysis uncovered active signaling pathways and potential therapeutic targets that were not evident from DNA or RNA data alone.
Methods
- Model Panel: 24 PDX models representing major breast cancer subtypes.
- Proteomics: Mass spectrometry‑based quantitative proteomics using both isobaric mass‑tag labeling and label‑free methods to enhance depth and cross‑validation of proteome coverage.
- Integration: Proteomic data were integrated with transcriptomic profiles to analyze correspondences and discordances between mRNA and protein levels.
- Analyses: Differential expression, subtype clustering, and pathway enrichment were conducted to identify biological signatures. Selected models were evaluated in drug treatment experiments to validate predictions.
Results
- Proteogenomic analysis validated multiple predicted genomic targets within receptor tyrosine kinase pathways, such as ERBB2/HER2.
- Several protein and phosphoprotein regulatory events were identified that were not predictable from genomics alone.
- In in vivo xenograft treatment experiments, responses to HER2‑targeted agents and PI3K pathway inhibition were consistent with proteogenomic signature predictions, supporting the idea that proteomic features may inform therapeutic sensitivity.
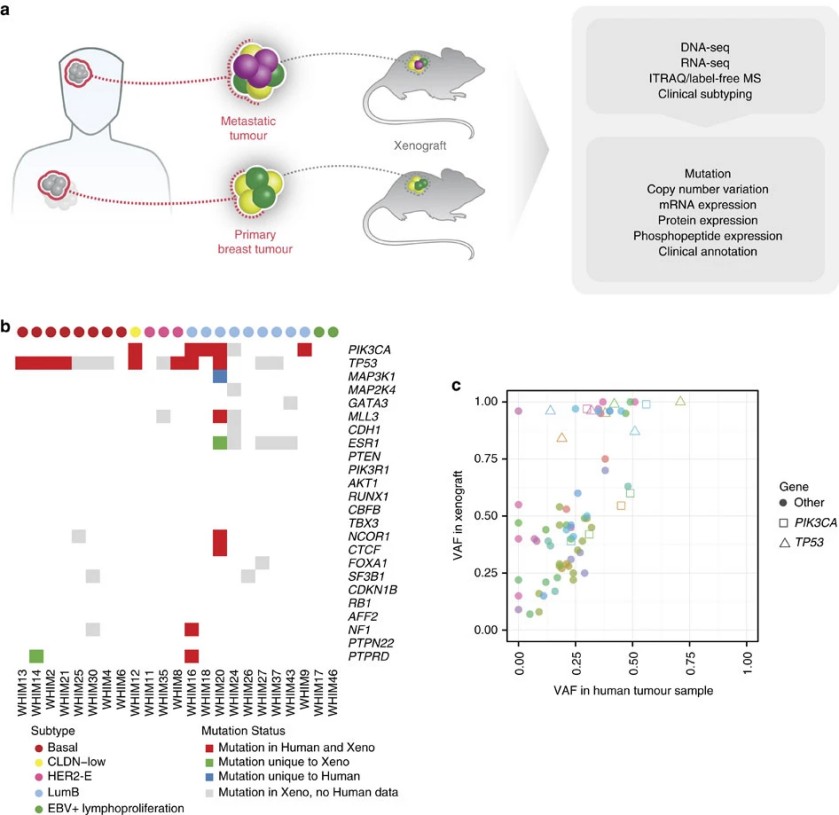
Figure 4. Modelling human breast cancer with patient-derived xenografts.
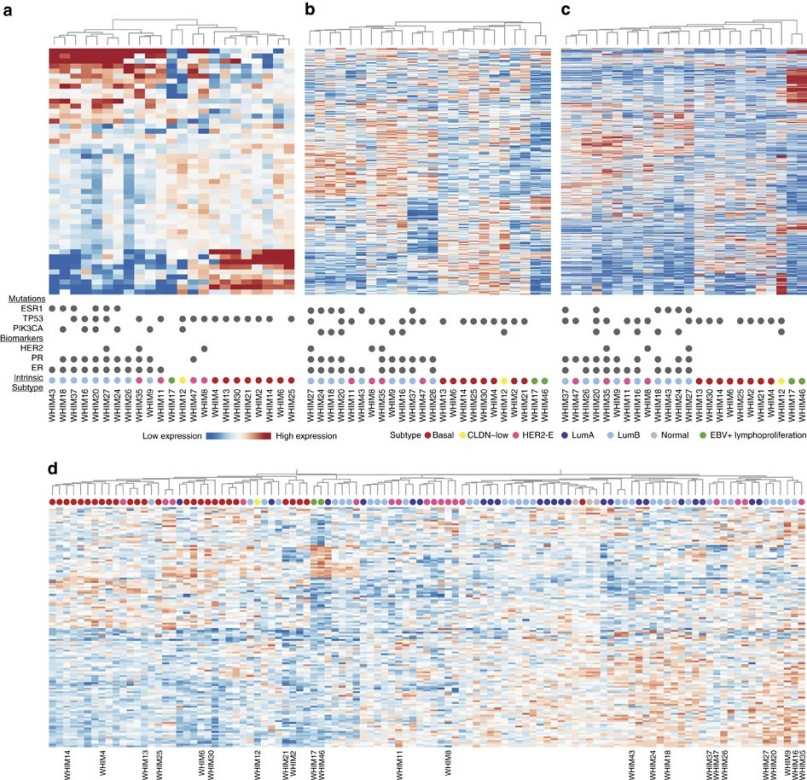
Figure 5. Transcriptomic and proteomic clustering of breast cancer PDX and human samples.
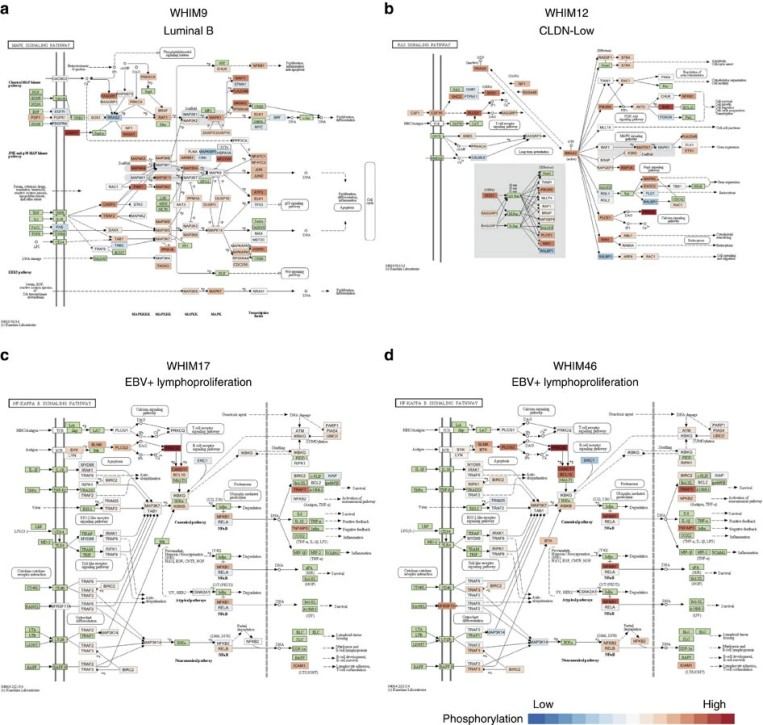
Figure 6. Activated signalling pathways detected through pathway phosphorylation enrichment analysis.
Conclusion
This study shows that combining proteomics with genomic data in PDX models provides a more complete view of cancer biology than DNA or RNA analysis alone. Mass spectrometry-based proteomics reveals changes in protein activity and signaling that are not captured at the gene level.
Related Services
References
- Liu Y, et al. Patient-derived xenograft models in cancer therapy: technologies and applications. Signal Transduction and targeted therapy, 2023, 8(1): 160.
- Uzozie A C, et al. PDX models reflect the proteome landscape of pediatric acute lymphoblastic leukemia but divert in select pathways. Journal of Experimental & Clinical Cancer Research, 2021, 40(1): 96.
- Huang K, et al. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nature communications, 2017, 8(1): 14864.