Drug Discovery & Translational Proteomics
Effective drug development begins with identifying the right targets, understanding their mechanism of action, and validating compound engagement in relevant biological systems. Creative Proteomics offers a full-spectrum proteomics workflow, enabling researchers to move seamlessly from discovery to translational validation
Integrated Proteomics Solutions for Drug Discovery & Translational Research
Despite unprecedented investment in pharmaceutical R&D, drug discovery continues to face a fundamental challenge: high attrition rates driven by insufficient molecular validation. Many drug candidates advance into costly preclinical or early clinical research phases without robust confirmation of target engagement, mechanism of action, or translational relevance. As a result, late-stage failure often reflects not poor chemistry, but gaps in biological evidence generated earlier in the pipeline.
Proteomics has become an essential tool in drug discovery. By directly measuring protein levels, interactions, modifications, and stability, mass spectrometry-based proteomics offers system-level insights that genomics or transcriptomics alone cannot provide. Strategically applied, it allows researchers to verify that a compound engages its intended target in live cells, detect unintended off-target effects, identify pharmacodynamic biomarkers, and evaluate how well preclinical models reflect human biology.
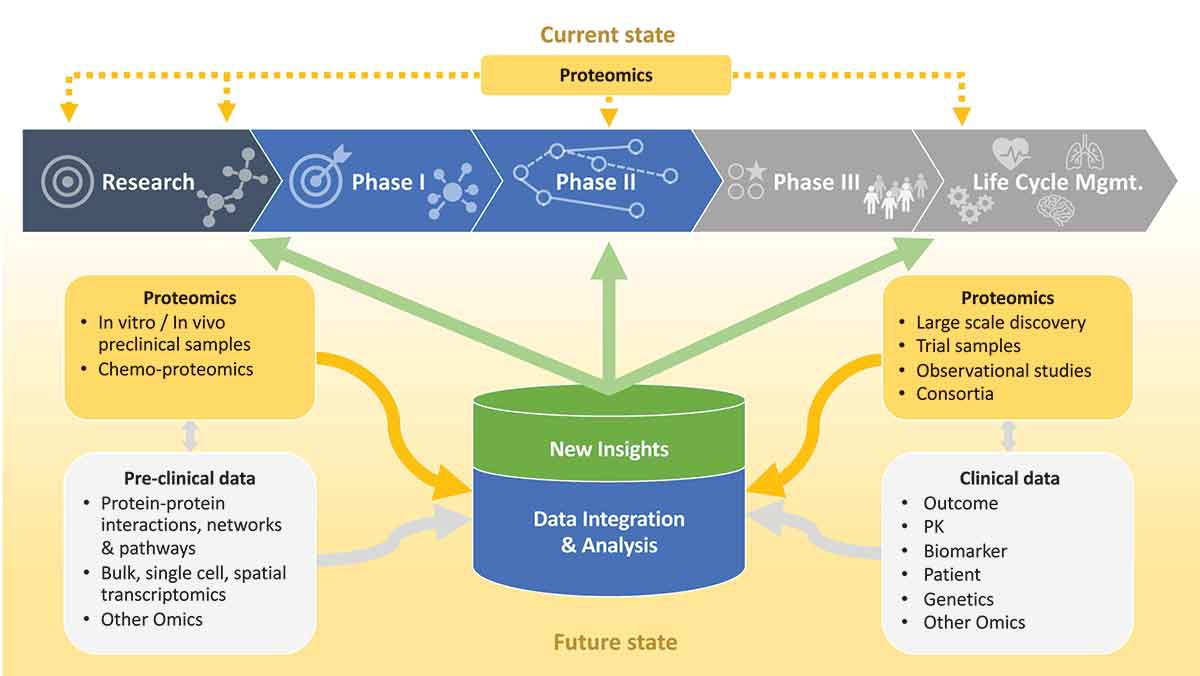
Figure 1. Current and future state of proteomics in the pharmaceutical industry (Lill J R, et al., 2021).
Creative Proteomics provides integrated proteomics solutions for Drug Discovery and Translational Research, connecting early target discovery to downstream clinical research. Our workflows generate reliable, decision-ready proteomic data to support pharmaceutical and biotech R&D teams across discovery, preclinical assessment, and biomarker development. Leveraging advanced mass spectrometry and validated bioinformatics pipelines, we help researchers de-risk drug programs, explore challenging targets, and improve the accuracy of preclinical data.
Target Discovery & Validation
Drug Target Deconvolution via Thermal Proteome Profiling (TPP)
Problem: In early drug discovery, demonstrating biochemical activity against a purified protein is insufficient. Compounds may fail to engage their targets in living cells due to permeability issues, protein complexation, or compensatory biology. Without direct evidence of cellular target engagement, programs risk advancing false positives into costly downstream studies.
Solution: TPP is a label-free, mass spectrometry-based method that directly measures ligand-induced changes in protein thermal stability across the proteome. When a compound binds to a protein, either directly or indirectly through complex formation, it alters the protein’s resistance to thermal denaturation. In a typical TPP workflow, intact cells or lysates are treated with a compound and exposed to a temperature gradient. Soluble proteins are quantified using high-resolution LC-MS/MS, generating melting curves for thousands of proteins simultaneously. Shifts in melting temperature (ΔTm) provide direct evidence of target engagement and allow systematic interrogation of the mechanism of action.
Covalent Drug Targeting and Cysteine Profiling
Problem: Many disease-driving proteins are considered "undruggable" because they lack typical binding pockets. Covalent inhibitors can target these proteins, but unintended reactivity may cause toxicity or misleading results.
Solution: Cysteine profiling uses chemical probes and quantitative proteomics to identify reactive cysteine residues in proteins under near-physiological conditions. By pinpointing hyper-reactive, ligandable cysteines, this method guides the design of covalent inhibitors with higher selectivity. Competitive labeling experiments reveal which cysteines interact with a test compound and assess off-target effects across the proteome, supporting safer and more effective drug design.
Problem: Targeted protein degradation has revolutionized drug discovery, but degrader molecules can create new challenges. It is critical to confirm that the degrader selectively eliminates the intended protein, triggers proper ubiquitination, and does not disrupt other proteins, ensuring reliable evaluation of its effectiveness.
Solution: We perform global proteome analysis to track protein degradation and ubiquitination across the cell. By measuring protein turnover on a proteome-wide scale, researchers can confirm that degraders engage their intended targets, assess selectivity, and detect unintended degradation of off-target proteins. This strategy helps reduce risks in developing next-generation therapies for proteins that were previously considered difficult to target.
Clinical & Preclinical Translation
Problem: Genomic predictions of neoantigens often overestimate which peptides are actually displayed on tumor cells. Direct evidence is critical to avoid pursuing non-immunogenic targets in immunotherapy research.
Solution: We use deep immunopeptidomics to pull HLA-bound peptides directly from tumor samples, cell lines, or patient-derived xenografts and identify them by high-resolution mass spectrometry. Because these peptides are observed on the cell surface, they provide biologically validated targets for immunotherapy and cancer vaccine research. Our bioinformatics maps each peptide to the tumor mutations and estimates immune relevance to prioritize candidates for translational studies.
Biomarker Discovery & Verification
Problem: Biomarkers discovered in small cohorts often fail to reproduce due to technical variability or biological noise. Translational success requires a structured pipeline from discovery to verification.
Solution: We provide complete Biomarker R&D Services, beginning with unbiased discovery using DIA proteomics to capture wide-ranging protein changes across samples. Promising candidates are then validated with targeted approaches like PRM or other quantitative mass spectrometry methods. This structured workflow allows confident selection of biomarkers for translational studies, preclinical research, and support of companion diagnostic development.
PDX/CDX Model Characterization
Problem: PDX and CDX are widely used for preclinical efficacy assessment, but their predictive value depends on how well they mirror human tumor proteomes and microenvironment features. Unrecognized species-specific signals or stromal replacements can mislead efficacy interpretation and degrade preclinical data fidelity.
Solution: Our PDX/CDX model characterization service uses quantitative proteomics to assess the molecular fidelity of preclinical models relative to human tissues. By comparing signaling pathways, protein expression patterns, and disease-relevant networks, we help determine whether a model is truly predictive of human response. This proteome-level validation supports informed model selection, improves translational relevance, and strengthens confidence in preclinical efficacy data used to guide investigational studies.
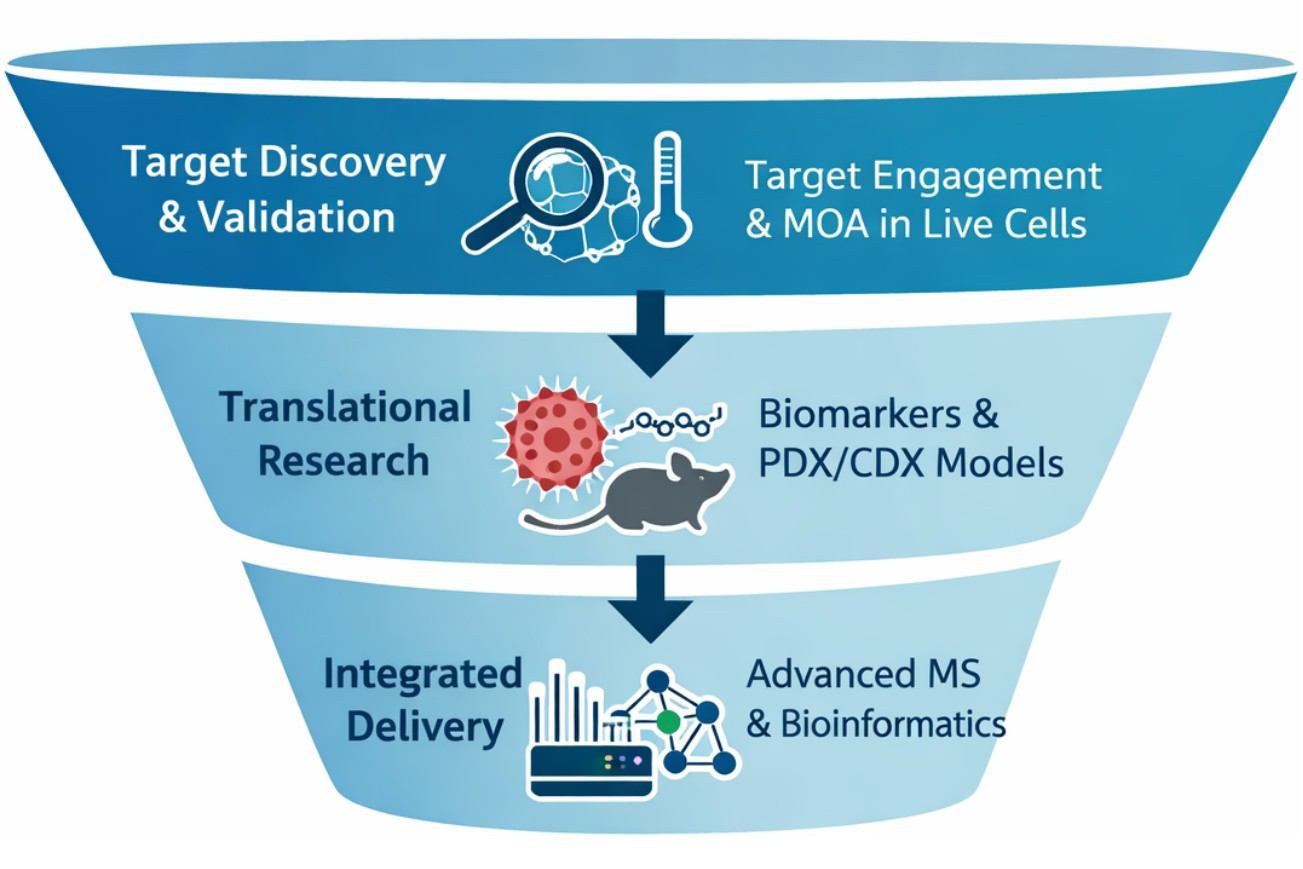
Workflow of Drug Discovery & Translational Proteomics
Successful drug development requires a streamlined workflow that ensures high-fidelity data and actionable insights at every stage. Our approach spans the entire drug discovery funnel:
- Target Discovery & Validation (Discovery Phase): We deconvolute targets and verify target engagement and mechanism of action in live cells using orthogonal proteomics approaches (label-free TPP, cysteine reactivity profiling, and ubiquitin/turnover profiling for degraders).
- Translational Research (Translational Phase): We identify HLA-presented neoantigens and immunopeptidomes, run discovery-to-verification biomarker pipelines (DIA to PRM), and perform PDX/CDX model characterization to ensure preclinical data fidelity.
- Integrated Delivery: Our integrated approach combines high-resolution mass spectrometry with advanced bioinformatics, including pathway analysis, protein–protein interaction (PPI) networks, and mechanism of action clustering, to generate insights that inform experimental design, compound prioritization, and translational strategy.
Our Services
Our Advanced Technology Platform
- Orbitrap Hybrid MS Systems: High-resolution and accurate mass detection allow precise identification and measurement of proteins in complex samples. This reliability is essential for TPP and PRM.
- timsTOF/4D-Proteomics: By combining trapped ion mobility (TIMS) with time-of-flight detection, timsTOF adds an extra dimension of separation. This improves protein identification and increases coverage, making it ideal for studying HLA-bound peptides and low-abundance neoantigens from limited samples.
- Nano-LC Systems: Using low-flow rates improves peptide separation and boosts sensitivity, making it easier to detect low-abundance biomarkers and HLA-presented peptides.
- Enrichment Hardware & Automation: For ubiquitin remnant enrichment, HLA immunoprecipitation, and cysteine probe chemistries, we use validated automated platforms to improve throughput and reproducibility.
Integrated Bioinformatics Analysis for Drug Discovery and Translation
Delivering mass spectrometry data is necessary but insufficient for decision-grade insight. Our bioinformatics layer transforms proteomic outputs into biological narratives you can act on.
- Functional Enrichment: We perform KEGG and Gene Ontology (GO) enrichment to highlight perturbed pathways and biological processes linked to compound exposure or model differences.
- Protein-Protein Interaction (PPI) Networks: Network analysis identifies key hub proteins and groups of co-regulated proteins, revealing potential backup pathways. Mapping PPIs places targets within cellular networks and helps anticipate downstream effects or guide combination strategies.
- Mechanism of Action Clustering & Signature Matching: By clustering proteome signatures across compounds or conditions, we can infer mechanism of action or group compounds with similar biological effects—useful for repurposing or lead optimization.
- Quantitative Reporting & Statistical Rigor: We implement multiple-comparison correction, estimate effect sizes, and use resampling when justified to control false discoveries and quantify confidence. Final outputs include validated, machine-readable files and summary tables designed for direct import into data lakes and downstream modeling pipelines.
- Deliverables: Interactive reports, volcano plots, pathway diagrams, prioritized candidate lists, and machine-readable results for downstream modeling or integration with genomics.
Why Partner With Us?
- Pharma-Grade Standards: Validated workflows, sample-tracking SOPs, and batch QC metrics to maximize reproducibility.
- Advanced Tech Stack: Access to leading MS platforms (Orbitrap, timsTOF) and enrichment chemistries for specialized assays.
- Integrated Interpretation: Biological insights delivered by experienced scientists with domain knowledge in drug discovery and translational research.
- Risk Mitigation: Services explicitly designed to de-risk drug pipelines by resolving target engagement ambiguity, exposing off-target liabilities, and improving preclinical data fidelity.
Frequently Asked Questions (FAQ)
-
Q1: What level of data and interpretation is delivered at project completion?
A1: Deliverables typically include processed quantitative datasets, detailed bioinformatics analyses (GO, KEGG, PPI networks, mechanism of action clustering), and an interpretive report highlighting key biological findings. This ensures the data is immediately usable for internal R&D discussions and downstream experimental planning.
-
Q2: How do proteomics data integrate with other omics or screening datasets?
A2: Proteomics data can be integrated with genomics, transcriptomics, CRISPR screens, or phenotypic assays to build a more comprehensive biological model. Our bioinformatics workflows support cross-omics interpretation, enabling pathway-level insights and more robust hypothesis generation in translational research.
-
Q3: How do label-free and labeled (TMT) approaches differ?
A3: Label-free quantification enables direct comparison of protein abundance across conditions without chemical labeling, while TMT allows multiplexed analysis for increased throughput and precision.
-
Q4: Are low-input or scarce samples suitable for these workflows?
A4: Many of our workflows are optimized for limited sample amounts. High-sensitivity platforms such as timsTOF and Orbitrap, combined with Nano-LC, enable deep proteome and immunopeptidome coverage from scarce or precious research samples, including PDX tissues.
-
Q5: How do you ensure off-target effects are accurately captured?
A5: We combine global proteome analysis, covalent cysteine profiling, and TPP to detect off-target effects and protein degradation, ensuring reliable preclinical data.
-
Q6: How early in a drug discovery program can proteomics be applied?
A6: Proteomics can be integrated at very early stages, including hit-to-lead and lead optimization. Techniques such as TPP, covalent cysteine profiling, and global proteome analysis help confirm target engagement, assess mechanism of action, and flag off-target effects before advancing to in vivo studies, thereby helping to de-risk the drug pipeline.
Creative Proteomics is positioned to support your discovery-to-translation journey with Integrated Proteomics Solutions that provide decision-grade evidence and reduce program risk. If you are interested, please contact us for more information and a quote.
Reference
- Lill J R, et al. Proteomics in the pharmaceutical and biotechnology industry: a look to the next decade. Expert Review of Proteomics, 2021, 18(7): 503-526.

