Thermal Proteome Profiling (TPP) Service
What is Thermal Proteome Profiling (TPP)?
Thermal Proteome Profiling (TPP) is a proteomics approach that links ligand or compound binding to changes in protein thermal stability. Proteins exhibit characteristic melting profiles, and when bound by a small molecule or metabolite, their thermal stability may increase or decrease. By measuring protein solubility across a temperature gradient, TPP allows unbiased identification of on-targets, off-targets, and mechanistic changes in native cellular and tissue environments. Thermal proteome profiling enables unbiased, proteome-wide identification of protein target engagement and pathway-level effects, bridging phenotypic observations with molecular mechanisms in drug discovery & and translational proteomics
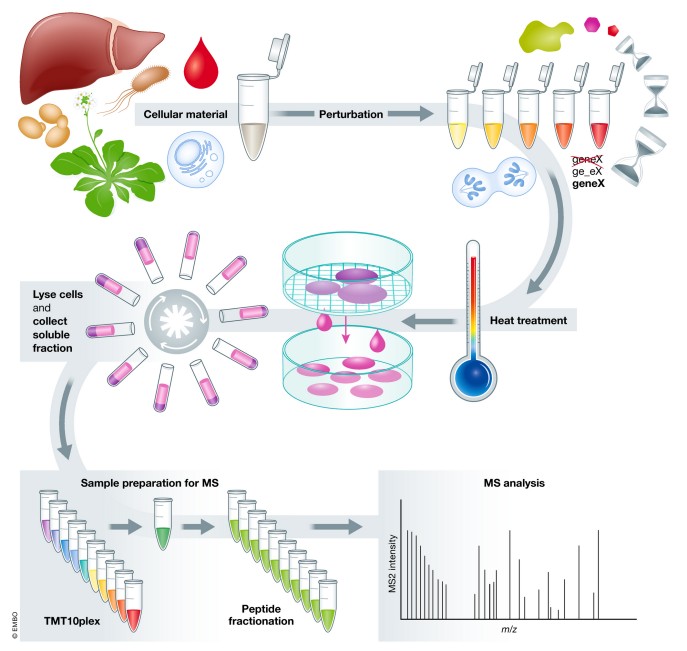
Figure 1. Thermal proteome profiling (TPP) experimental setup (Mateus A, et al., 2020).
Advantages of TPP for Drug Target Identification
- Unbiased, Proteome-Wide Coverage: TPP monitors thousands of proteins simultaneously, capturing both expected targets and off-target interactions.
- Probe-Free and Label-Free: Compounds remain unmodified, preserving their intrinsic properties and biological activity.
- Direct and Indirect Effects: Beyond primary binding events, TPP detects protein complex changes, post-translational modifications, and pathway rewiring.
- Quantitative and High-Resolution: Coupled with LC–MS/MS, TPP provides precise, reproducible measurements for data-driven target prioritization.
Data Analysis and Interpretation
- Melting Curve Modeling: Tracks protein abundance across temperatures to identify stability shifts.
- Nonparametric Analysis of Response Curves (NPARC): Prioritizes proteins based on the overall denaturation profile rather than a single melting point.
- Candidate Prioritization: Combines thermal stability data with functional annotation, transcriptomics, or other proteomic datasets to identify biologically meaningful effectors.
Complementary Techniques and Multi-Omics Integration
TPP results can be integrated with other proteomics and omics approaches to provide a holistic understanding:
- Transcriptomics Integration: Confirms changes in gene expression corresponding to protein stability alterations.
- PTM Analysis: Identifies post-translational modifications affecting protein function and stability.
- Protein-Protein Interaction Studies: Reveals complex formation or dissociation events linked to compound treatment.
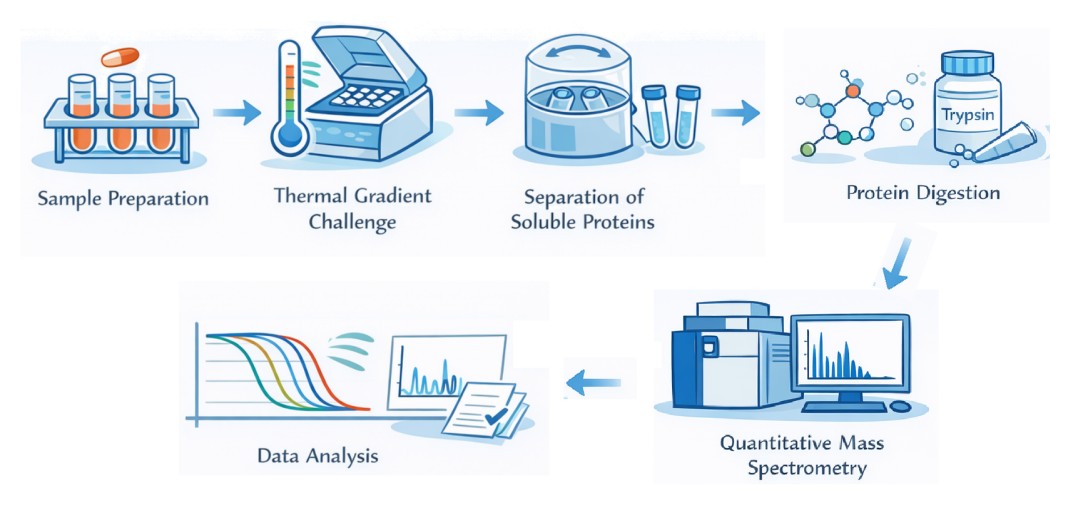
How Thermal Proteome Profiling Works?
- Sample Preparation: Cells or tissues are treated with the compound of interest.
- Thermal Gradient Challenge: Samples are briefly heated across a temperature range to induce protein unfolding.
- Separation of Soluble Proteins: Aggregated proteins are removed via centrifugation, leaving soluble, non-denatured proteins.
- Protein Digestion: The remaining proteins are enzymatically digested into peptides, typically using trypsin.
- Quantitative Mass Spectrometry: Peptides are labeled with isobaric tags and analyzed by LC-MS/MS.
- Data Analysis: Protein abundance is modeled as a function of temperature to generate melting curves, identifying proteins whose stability is affected by compound treatment.
Deliverables and Reporting Standards
- Raw LC–MS/MS spectra
- Quantified peptide and protein abundance tables
- Protein melting curves and prioritized hit lists
- Functional and pathway annotations
- Interactive visualizations and recommended follow-up strategies
Applications of TPP in Biomedical Research
- Small Molecule Target Discovery: Identifies potential protein targets of new compounds.
- Off-Target Profiling: Detects unintended interactions to inform mechanistic understanding.
- Pathway Mapping: Reveals downstream signaling and protein complex changes.
- Drug Resistance Studies: Evaluates proteins contributing to cellular adaptation under compound treatment.
- Natural Product and Metabolite Studies: Characterizes biological targets of metabolites or bioactive compounds.
- Functional Annotation: Provides insights into the role of protein targets within cellular processes.
Sample Requirements
| Sample type | Recommended input (typical) | Handling & storage |
| Adherent cell lines | 1–5 × 107 cells per condition | Keep on ice after harvest, snap-freeze pellets if not processed immediately; avoid repeated freeze–thaw. |
| Suspension cell lines | 1–5 × 107 cells per condition | Pellet cells quickly at 4°C, freeze pellets on dry ice if needed. |
| Fresh or frozen tissues | ≥50 mg fresh tissue or equivalent frozen material per condition | Store at −80°C; keep tissue frozen until lysis. |
| Cell/tissue lysates | ≥50–100 µg total protein per temperature point | Keep on ice during processing; aliquot and freeze at −80°C if not immediately digested. |
| Purified proteins / recombinant enzymes | 1–10 µg per assay point | Store per protein specifications; avoid detergents that affect thermal behavior. |
Why Choose Creative Proteomics for TPP?
Creative Proteomics provides comprehensive TPP services, backed by over 20 years of expertise in quantitative proteomics. Key advantages include:
- High-resolution LC-MS/MS platforms capable of deep proteome coverage.
- Optimized sample preparation and workflow options for diverse biological systems.
- Reproducible, high-confidence datasets ready for downstream analysis.
- Customizable study designs to address specific research questions or experimental constraints.
FAQ
-
Q1: What formats of TPP experiments are commonly used?
A1: Common TPP formats include temperature range experiments (TPP-TR), concentration range experiments (TPP-CCR), and combined two-dimensional experiments (2D-TPP).
-
Q2: Is TPP probe‑free and label‑free?
A2: Yes, TPP itself does not require chemical modification of compounds (probe‑free), though it typically uses multiplexing labels (e.g., TMT) for quantitative mass spectrometry.
-
Q3: What factors influence TPP results?
A3: Temperature range selection, compound concentration, sample type, protein abundance, and data processing methods all affect the detection of thermal shifts.
-
Q4: What are common limitations of TPP?
A4: Limitations include reliance on detectable protein abundance, complexity of data analysis, and challenges in analyzing membrane proteins without solubilization strategies.
-
Q5: How does TPP compare with other target identification methods?
A5: Unlike affinity purification that uses probes or tags, TPP detects physical consequences of ligand binding on protein stability, offering unbiased proteome‑wide insights without prior modification of compounds.
Demo
Demo: Thermal proteome profiling reveals fructose-1,6-bisphosphate as a phosphate donor to activate phosphoglycerate mutase 1
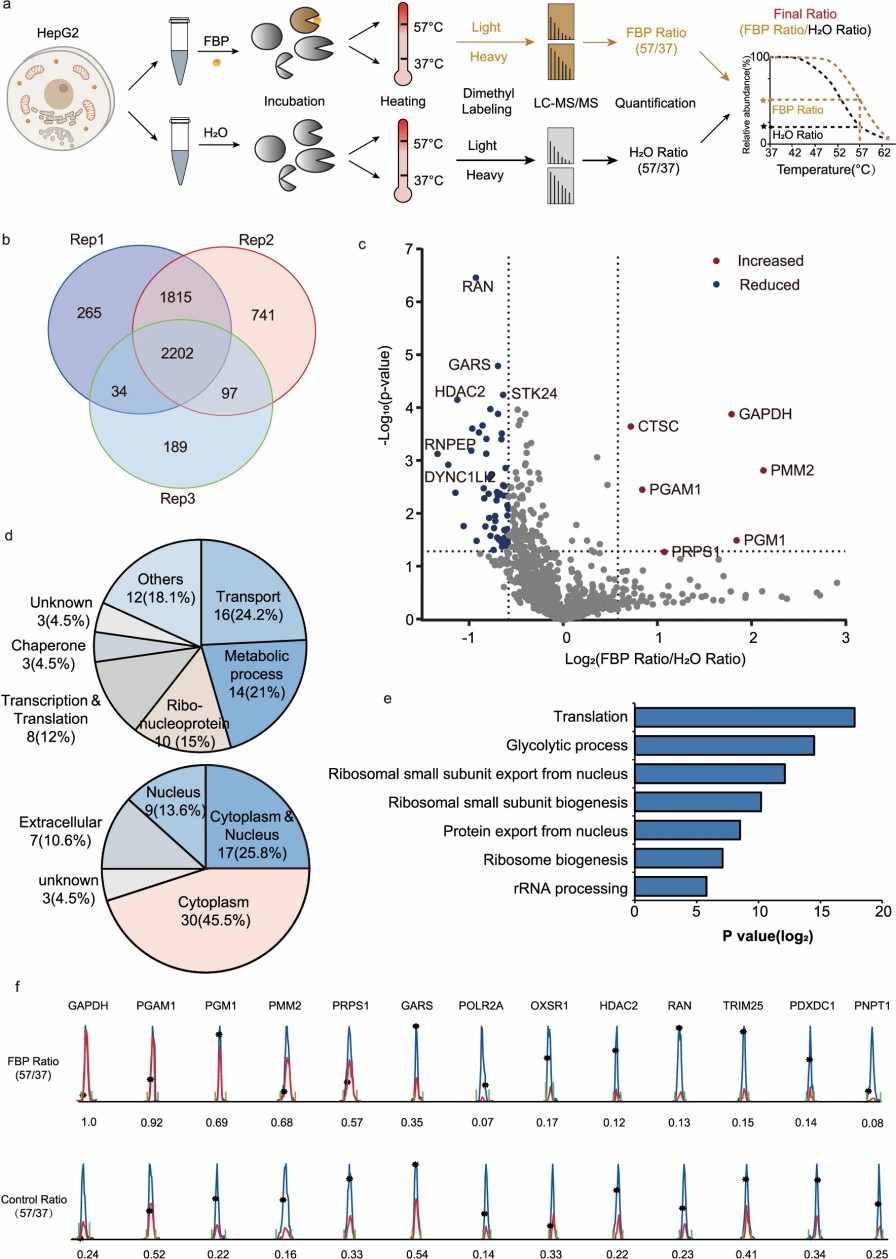
Figure 2. Thermal proteome profiling (TPP) of FBP-interacting proteins (Zhang Y, et al., 2024).
-
Case Study
Case: Characterization of a small molecule inhibitor of disulfide reductases that induces oxidative stress and lethality in lung cancer cells
Abstract
The primary aim was to identify the protein targets responsible for the selective cytotoxicity of a small molecule (referred to in the paper as a hit from a lung cancer screen) toward lung adenocarcinoma cells, and to validate whether those targets explain the observed induction of oxidative stress and cell death. The authors intended to show that TPP can discover biologically meaningful effectors from a phenotypic hit and to validate the biochemical and cellular consequences of target engagement.
Methods
- Applied thermal proteome profiling (TPP) on treated cancer cell lysates/cells to detect protein thermal-stability shifts after LCS3 exposure.
- Quantitative mass spectrometry (TMT/LC–MS/MS) reconstructed melting curves and prioritized hits.
- Follow-up validation included purified-enzyme activity assays, functional cellular assays, and a genome-wide CRISPR screen to link targets to sensitivity.
Results
- TPP nominated the disulfide reductases GSR and TXNRD1 as top targets.
- Biochemical assays confirmed LCS3 inhibits these enzymes; treated cells showed increased reactive-oxygen-species signaling and an NRF2-like oxidative-stress response.
- A CRISPR screen identified genetic modifiers that modulate sensitivity, supporting the biological relevance of the identified targets.
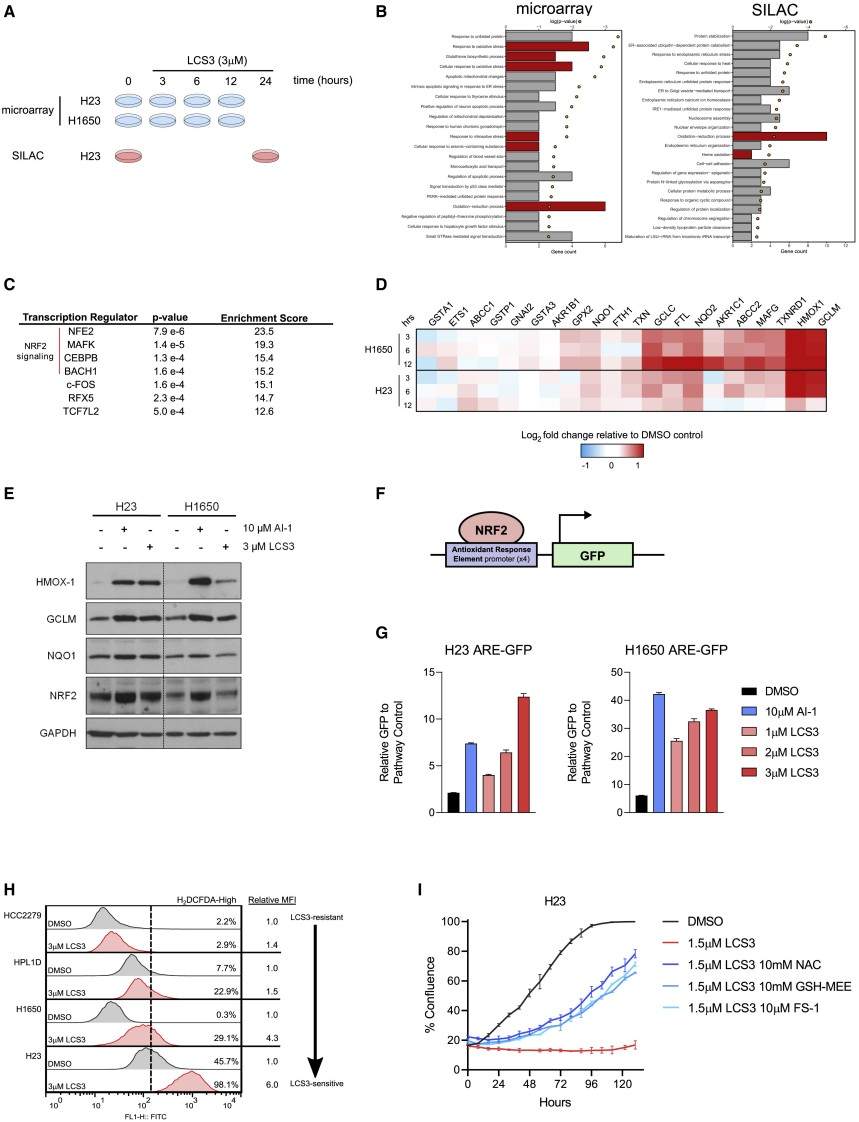
Figure 3. LCS3 induces ROS and NRF2 pathway activation.
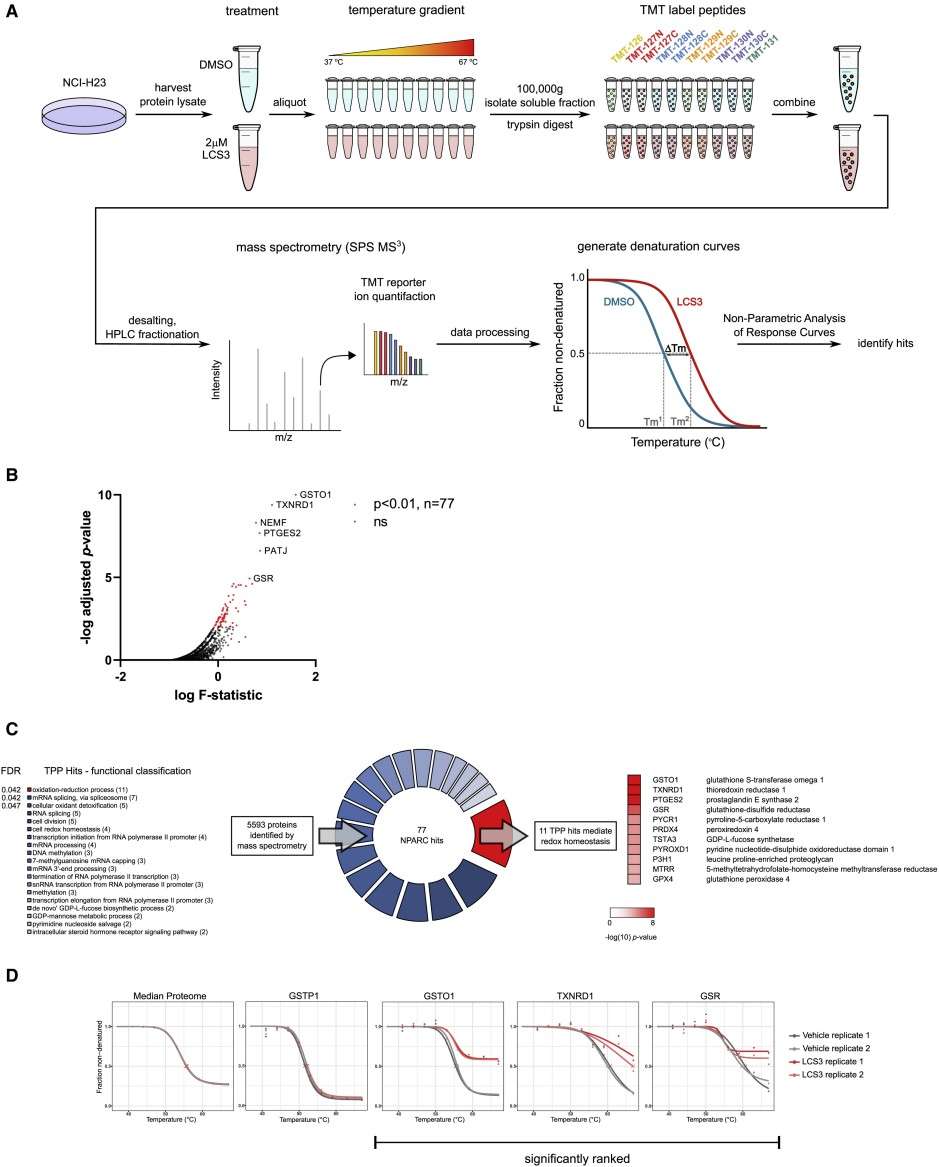
Figure 4. TPP identifies candidate LCS3-interacting proteins that mediate redox homeostasis.
Conclusion
TPP successfully deconvoluted the compound's mechanism: LCS3 directly engages and inhibits GSR and TXNRD1, disrupting cellular redox buffering and causing selective cytotoxicity in susceptible lung adenocarcinoma cells. Orthogonal biochemical and genetic validation established these proteins as functionally meaningful effectors of the phenotype.
Related Services
References
- Mateus A, et al. Thermal proteome profiling for interrogating protein interactions. Molecular systems biology, 2020, 16(3): e9232.
- Zhang Y, et al. Thermal proteome profiling reveals fructose-1, 6-bisphosphate as a phosphate donor to activate phosphoglycerate mutase 1. Nature Communications, 2024, 15(1): 8936.
- Johnson F D, et al. Characterization of a small molecule inhibitor of disulfide reductases that induces oxidative stress and lethality in lung cancer cells. Cell reports, 2022, 38(6).