ICH Topic Q6B states: "If, based on the gene sequence for the desired product, cysteine residues are expected, the number and positions of any free sulfhydryl groups and/or disulfide bridges should be determined, to the extent possible. Peptide mapping (under reducing and non-reducing conditions), mass-spectrometry, or other appropriate techniques may be useful for this evaluation."
At Creative Proteomics, we are committed to offering comprehensive disulfide bridge (S-S) analysis service to determine understand protein structure and function, reveal protein stability, discover protein interactions, and optimize protein modifications.
Why do Disulfide Bridge (S-S) Analysis?
Disulfide bridges, often represented as S-S bonds, are covalent bonds formed between two sulfur atoms in different cysteine residues within a protein. Cysteine is an amino acid that contains a thiol group (-SH) on its side chain. When two cysteine residues come into close proximity, oxidation can occur, leading to the formation of a disulfide bond.
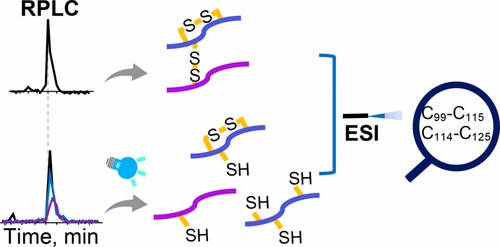 Fig 1. Mapping Complex Disulfide Bonds via Implementing Photochemical Reduction Online with Liquid Chromatography−Mass Spectrometry. (Yang, X., & Xia, Y. 2020)
Fig 1. Mapping Complex Disulfide Bonds via Implementing Photochemical Reduction Online with Liquid Chromatography−Mass Spectrometry. (Yang, X., & Xia, Y. 2020)
The disulfide bridge plays a crucial role in stabilizing the three-dimensional structure of proteins. By forming covalent bonds between specific cysteine residues, disulfide bridges help to maintain the proper folding and stability of proteins.
What can we offer for disulfide bridge (S-S) analysis?
Ellman's Reagent Assay
This method utilizes 5,5'-dithiobis(2-nitrobenzoic acid) (DTNB), also known as Ellman's reagent, which reacts specifically with free sulfhydryl groups of reduced cysteine residues. By comparing the concentration of free sulfhydryl groups before and after reduction with a reducing agent like dithiothreitol (DTT), the number of disulfide bonds can be determined.
Mass spectrometry
Mass spectrometry can provide accurate and sensitive information about the presence and location of disulfide bonds. Techniques like MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization Time-of-Flight) and LC-MS/MS (Liquid Chromatography-Mass Spectrometry/Mass Spectrometry) can be used to analyze the intact protein or its enzymatic digests and identify the mass shifts caused by disulfide bond formation.
X-ray Crystallography
X-ray crystallography is a powerful technique to determine the three-dimensional structure of proteins. By growing protein crystals and analyzing the diffraction pattern of X-rays, the positions of the atoms including disulfide bonds can be determined.
NMR spectroscopy
Nuclear Magnetic Resonance (NMR) spectroscopy can provide structural information about proteins in solution, including the presence and locations of disulfide bonds. The observation of specific proton-proton and sulfur-proton interactions can be used to determine the connectivity of cysteine residues.
Gel electrophoresis (SDS-PAGE)
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) can be used to analyze the mobility of proteins in a gel matrix. Under reducing conditions, disulfide bonds are broken, resulting in a different mobility of reduced and non-reduced forms of the protein.
Applications of Disulfide Bridge
 Protein Folding and Stability
Protein Folding and Stability
 Protein Structure Determination
Protein Structure Determination
 Protein Engineering
Protein Engineering
 Drug Development
Drug Development
 Biomedical Research
Biomedical Research
Challenges in Identification of the Disulfide Bridges
- Tri- or even tetrapeptides that are disulfide linked and the size of the peptides.
- Posttranslational modifications
- Sample preparation
Sample Requirement
| Sample Type |
Protein |
Cell |
Animal tissue |
Plant Tissue |
Blood (EDTA added) |
Serum |
Urine |
Microbes |
| Quantity |
100 ug |
2X107 cells |
1g |
200 mg |
1mL |
0.2-0.5 mL |
2 mL |
Dry weighed: 200 mg |
Want to Learn More?
At Creative Proteomics, our primary goal is to provide our clients with exceptional disulfide bridge (S-S) analysis service that assist in the development of protein drugs. We continuously work to increase our knowledge and remain on the cutting edge of technological developments in protein characterization. We are dedicated to advancing the biopharmaceutical business and enhancing patient lives via our unwavering pursuit of service quality and effectiveness. If you are interested in our services or have any additional questions, please contact us for more information.
Reference
- Yang, X., & Xia, Y. Mapping Complex Disulfide Bonds via Implementing Photochemical Reduction Online with Liquid Chromatography–Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2020, 32(1), 307–314.
Related Sections
Services
Applications
For research use only, not intended for any clinical use.

 Fig 1. Mapping Complex Disulfide Bonds via Implementing Photochemical Reduction Online with Liquid Chromatography−Mass Spectrometry. (Yang, X., & Xia, Y. 2020)
Fig 1. Mapping Complex Disulfide Bonds via Implementing Photochemical Reduction Online with Liquid Chromatography−Mass Spectrometry. (Yang, X., & Xia, Y. 2020)




