ICH Topic Q6B states "Molecular weight (or size) is determined using size exclusion chromatography, SDS-polyacrylamide gel electrophoresis (under reducing and non-reducing conditions), mass-spectrometry, and other appropriate techniques".
Protein molecular weight analysis service provided by Creative Proteomics play an important role in studying the structure and function of proteins, as well as controlling the quality of protein products.
Protein Molecular Weight
Protein molecular weight pertains to the macromolecular mass of a protein molecule, typically quantified in units of Daltons (Da) or kilodaltons (kDa). This attribute is intrinsic to proteins and is influenced by the arrangement and type of constituent amino acids present in the protein's structure.
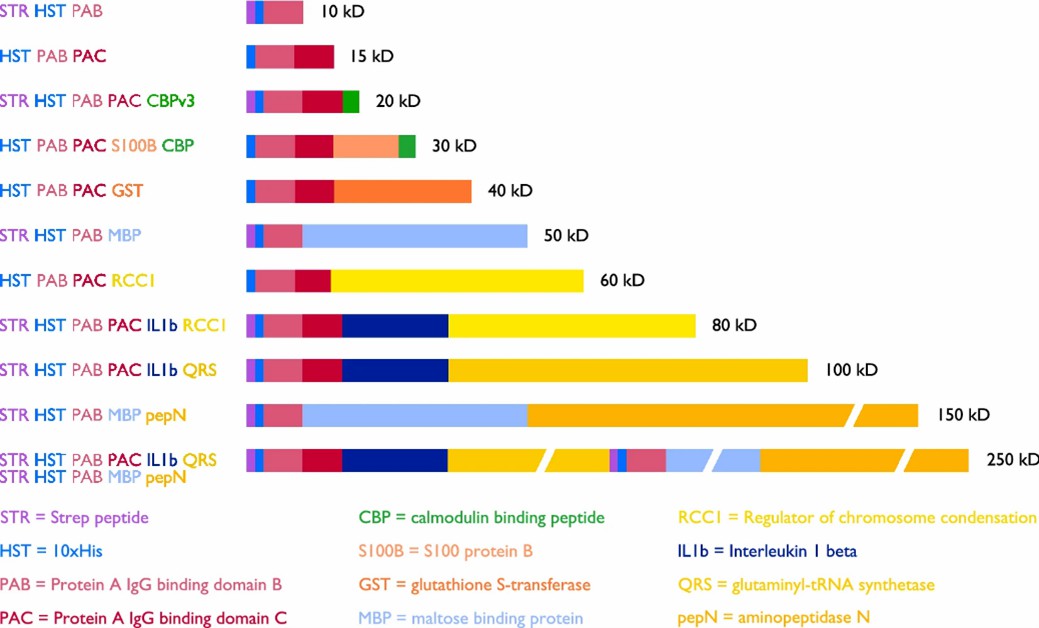 Fig 1. Schematic representation of Penn State ladder proteins with abbreviation key and color coding shown. (Santilli, R. T., et al.; 2021)
Fig 1. Schematic representation of Penn State ladder proteins with abbreviation key and color coding shown. (Santilli, R. T., et al.; 2021)
Applications of Protein Molecular Weight
Protein molecular weight is a crucial parameter in many research and application areas, including:
 Protein identification
Protein identification
Understanding a protein's molecular weight empowers researchers in verifying its identification and outlining its characteristics. By juxtaposing experimental molecular weight against predicted molecular weight derived from the protein sequence, scientists can affirm the identity of a protein and validate any post-translational modifications that may have occurred.
Protein purification 
Size-based separation techniques central to protein purification methods, like gel filtration chromatography, are reliant on molecular weight for their efficacy. Consequently, knowledge of the molecular weight of the target protein is essential for streamlining purification strategies and monitoring the success of the purification process.
 Protein structure analysis
Protein structure analysis
By evaluating and comparing the experimental and predicted molecular weights, it is possible to deduce the structural composition of the protein, determining whether the protein exists in the form of monomers, dimers, or higher oligomers. Such insight is invaluable for understanding protein functionality and possible interactions.
Biomarker discovery
Molecular weight characterization can also aid in the discovery of biomarkers – molecular indicators of the presence or development of specific diseases or conditions. Changes in the molecular weight of certain proteins could signal disease-related modifications or protein fragments.
 Protein-protein interactions
Protein-protein interactions
Knowledge of the molecular weights of interacting proteins is instrumental in the study of protein-protein interactions. This knowledge aids in comprehending the stoichiometry of complex formations, binding affinities between proteins, and the structural implications of such interactions.
Drug development
During the stages of drug discovery and development, measuring the molecular weight of a target protein is pivotal for the design and optimization of therapeutic agents. Ultimately, this enables researchers to assess the binding efficiency and specificity of potential drug candidates.
What can we offer?
There are several methods available for detecting protein molecular weight. The choice of method depends on factors such as the size and nature of the protein and the required level of accuracy. Here are some commonly used methods:
1. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
The technique in context employs an electric field as a vehicle for separating proteins using their molecular weight as the underlying principle. This is accomplished through the denaturation of proteins and subsequent coating with Sodium Dodecyl Sulfate (SDS). SDS functions by conferring a consistent negative charge on proteins, relative to their mass. This results in a separation of proteins within the gel as distinct bands, under electro-current, based solely on their relative sizes. Furthermore, for approximation of molecular weight, the migration patterns of these protein bands are juxtaposed with those of established molecular weight markers.
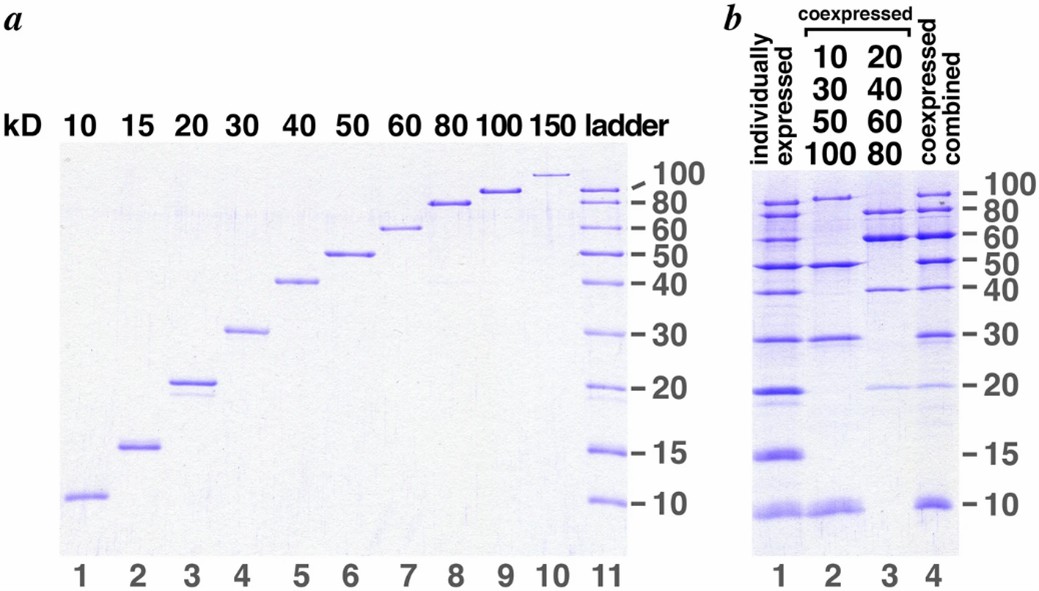 Fig 2. Protein molecular weight is detected by SDS-PAGE. (Santilli, R. T., et al.; 2021)
Fig 2. Protein molecular weight is detected by SDS-PAGE. (Santilli, R. T., et al.; 2021)
2. Western Blotting
In the post-electrophoretic process, proteins are segregated using SDS-PAGE, followed by their transference onto a nitrocellulose or polyvinylidene fluoride membrane, an operation known as Western blotting. Proteins that are the key focus are detected using specific antibodies, and through the interaction of these antibodies, the protein bands become illuminated and hence managed to be visualized. The molecular mass of these proteins is deducible via comparison of the band position in relation to previously calibrated molecular weight markers.
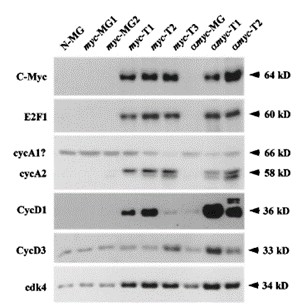 Fig 3. Western blot analyses for size comparisons of various proteins. (Liao, D. J., et al.; 2020)
Fig 3. Western blot analyses for size comparisons of various proteins. (Liao, D. J., et al.; 2020)
3. Mass Spectrometry
In this high-resolution technique, the protein is ionized and subjected to analysis based on the mass-to-charge ratio. The mass spectrometer can accurately determine the molecular weight of the protein by measuring the mass of the ions. This method is particularly useful for the analysis of large and complex proteins.
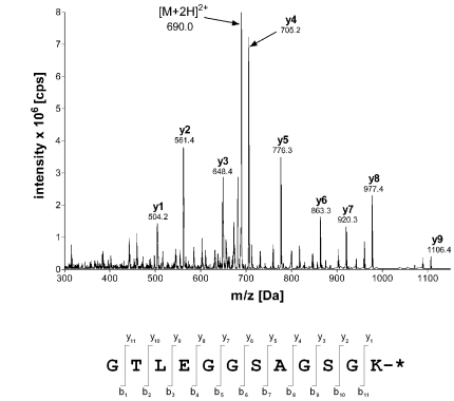 Fig 4. Mass spectrometry identification of the fluorescently labeled maltose binding protein ML. (Volkmann, G., & Liu, X.-Q.;2009)
Fig 4. Mass spectrometry identification of the fluorescently labeled maltose binding protein ML. (Volkmann, G., & Liu, X.-Q.;2009)
4. Gel Filtration Chromatography (Size Exclusion Chromatography)
This separation technique separates proteins based on their size using a column packed with porous beads. Smaller proteins enter the pores and have a longer retention time, while larger proteins pass freely through the column and have a shorter retention time. The molecular weight can be estimated by comparing the retention time of the protein to that of known molecular weight standards.
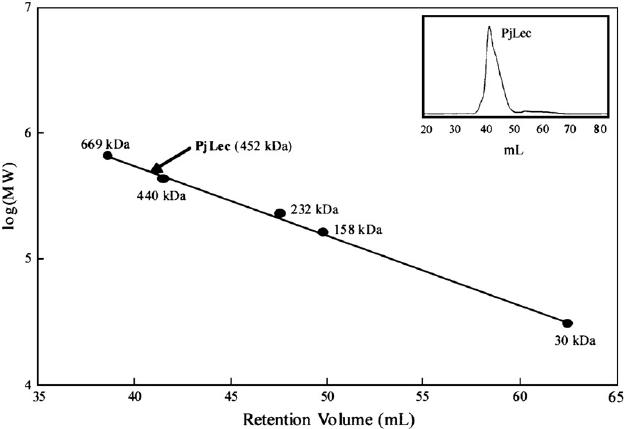 Fig 5. Gel filtration chromatography and determination of molecular size of PjLec. (YANG, H., et al.; 2007)
Fig 5. Gel filtration chromatography and determination of molecular size of PjLec. (YANG, H., et al.; 2007)
5. Analytical Ultracentrifugation
This method measures the sedimentation rate of proteins in a solution subjected to high centrifugal forces. The sedimentation coefficient is used to calculate the molecular weight of the protein. Analytical ultracentrifugation can analyze proteins ranging from small to large sizes.
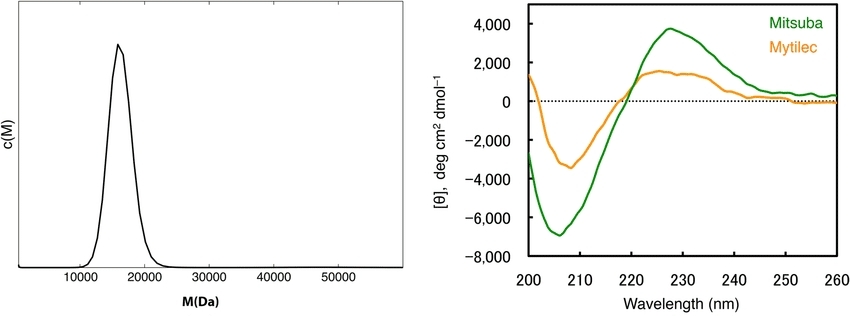 Fig 6. Molecular weight determination by analytical ultracentrifugation. (Terada, D., et al.; 2017)
Fig 6. Molecular weight determination by analytical ultracentrifugation. (Terada, D., et al.; 2017)
Our Advantages

Want to Learn More?
As a prominent biotechnological corporation, Creative Proteomics provides exhaustive protein molecular weight analysis services. These services are tailored to provide clients with meticulous and precise estimations of protein weights, utilizing state-of-the-art technology and progressive methodologies, all underpinned by rigorous analytical techniques. The objective of our services is to equip our clients with integral knowledge and comprehension about diverse proteins, a factor that can prove to be instrumental in their respective research or product development endeavors, propelling them towards considerable development and prosperity in their field. Please feel free to contact us.
References
- Santilli, R. T., et al.; The Penn State Protein Ladder system for inexpensive protein molecular weight markers. Scientific Reports. 2021, 11(1).
- Liao, D. J., et al.; Cell cycle basis for the onset and progression of c-Myc-induced, TGFα-enhanced mouse mammary gland carcinogenesis. Oncogene. 2020, 19(10), 1307–1317.
- Volkmann, G., & Liu, X.-Q. Protein C-Terminal Labeling and Biotinylation Using Synthetic Peptide and Split-Intein. PLoS ONE. 2009, 4(12), e8381.
- YANG, H., et al.; Purification and characterisation of a calcium-independent lectin (PjLec) from the haemolymph of the shrimp Penaeus japonicus. Fish & Shellfish Immunology. 2007, 22(1-2), 88–97.
- Terada, D., et al.; Computational design of a symmetrical β-trefoil lectin with cancer cell binding activity. Scientific Reports. 2017, 7(1).
Related Sections
Services
Applications
For research use only, not intended for any clinical use.

 Fig 1. Schematic representation of Penn State ladder proteins with abbreviation key and color coding shown. (Santilli, R. T., et al.; 2021)
Fig 1. Schematic representation of Penn State ladder proteins with abbreviation key and color coding shown. (Santilli, R. T., et al.; 2021) Protein identification
Protein identification
 Protein structure analysis
Protein structure analysis
 Protein-protein interactions
Protein-protein interactions
 Fig 2. Protein molecular weight is detected by SDS-PAGE. (Santilli, R. T., et al.; 2021)
Fig 2. Protein molecular weight is detected by SDS-PAGE. (Santilli, R. T., et al.; 2021) Fig 3. Western blot analyses for size comparisons of various proteins. (Liao, D. J., et al.; 2020)
Fig 3. Western blot analyses for size comparisons of various proteins. (Liao, D. J., et al.; 2020) Fig 4. Mass spectrometry identification of the fluorescently labeled maltose binding protein ML. (Volkmann, G., & Liu, X.-Q.;2009)
Fig 4. Mass spectrometry identification of the fluorescently labeled maltose binding protein ML. (Volkmann, G., & Liu, X.-Q.;2009) Fig 5. Gel filtration chromatography and determination of molecular size of PjLec. (YANG, H., et al.; 2007)
Fig 5. Gel filtration chromatography and determination of molecular size of PjLec. (YANG, H., et al.; 2007) Fig 6. Molecular weight determination by analytical ultracentrifugation. (Terada, D., et al.; 2017)
Fig 6. Molecular weight determination by analytical ultracentrifugation. (Terada, D., et al.; 2017)
